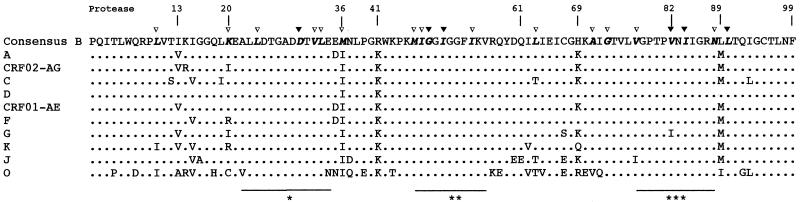
FIG. 2.
Amino acid alignment of protease consensus sequences for each subtype of group M and for group O. The sequences were aligned against a subtype B consensus sequence from the database; dots indicate homology. The amino acid positions associated with drug resistance are depicted in boldface italics; the major mutations (D30N, G48V, I50V, V82A/F/T, I84V, L90M) are marked at the top of the consensus sequence (▾), as are the minor mutations (L10I/V, K20M/R, L24I, V32I, L33F, M36I, M46I/L, I47V, I54L/V, L63P, A71T/V, G73S, V77I, N88D) (▿). The functional domains of the protease (∗, active site [amino acids 22 to 34]; ∗∗, flap region [amino acids 47 to 56]; ∗∗∗, substrate binding site [amino acids 78 to 88]) are shown under the consensus sequence.