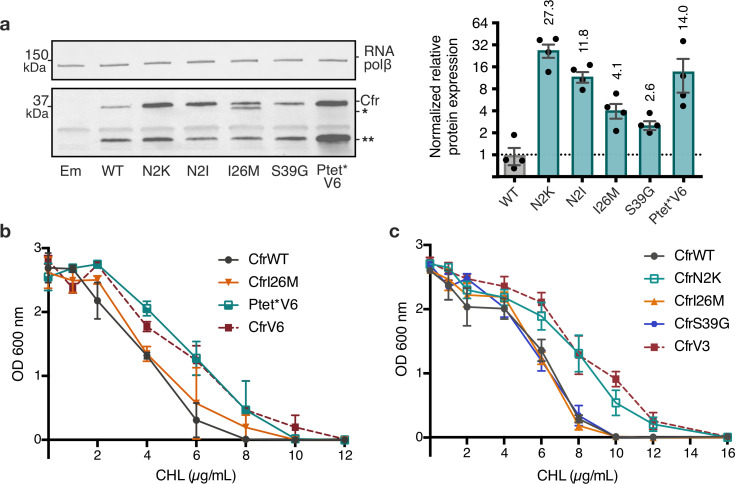
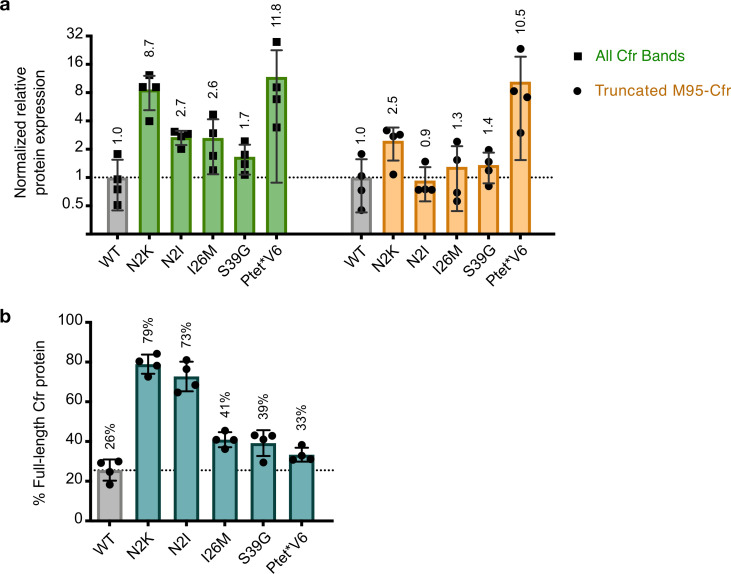
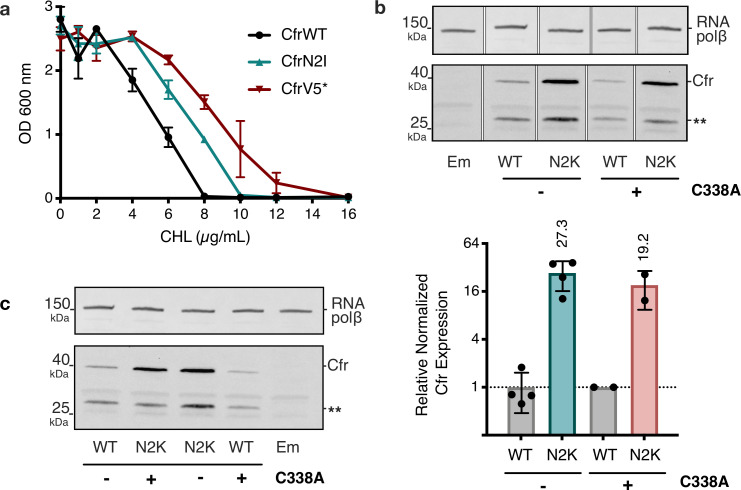
Figure 3. Mutations to the second amino acid and promoter are the largest contributors to Cfr expression and resistance.
(a) Effect of Cfr mutations and promoter alteration on relative Cfr protein expression was assessed by immunoblotting against a C-terminal FLAG tag. Quantification was performed for full-length Cfr protein normalized to housekeeping protein RNA polymerase β-subunit. Data are presented as the average of four biological replicates with standard deviation on a log2 axis. Asterisks denote N-terminally truncated Cfr protein products that do not contribute to resistance and were not included in quantification. Em = empty vector control. Original uncropped blot images are shown in Figure 3—source data 1. (b) and (c) Dose-dependent growth inhibition of Escherichia coli expressing pZA-encoded CfrWT, CfrV6 (panel (b)), CfrV3 (panel (c)), and individual mutants that comprise these variants toward chloramphenicol (CHL) presented as an average of three biological replicates with standard error. Numerical data for all figure panels are provided in Figure 3—source data 2.