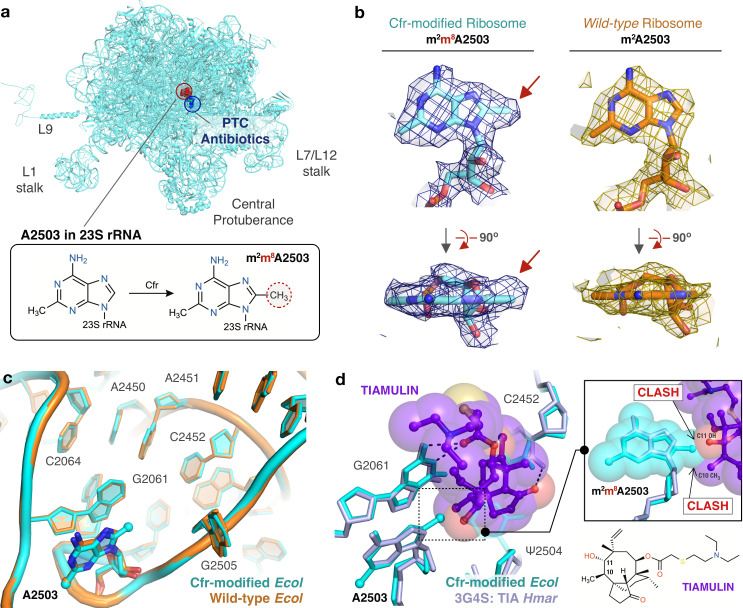
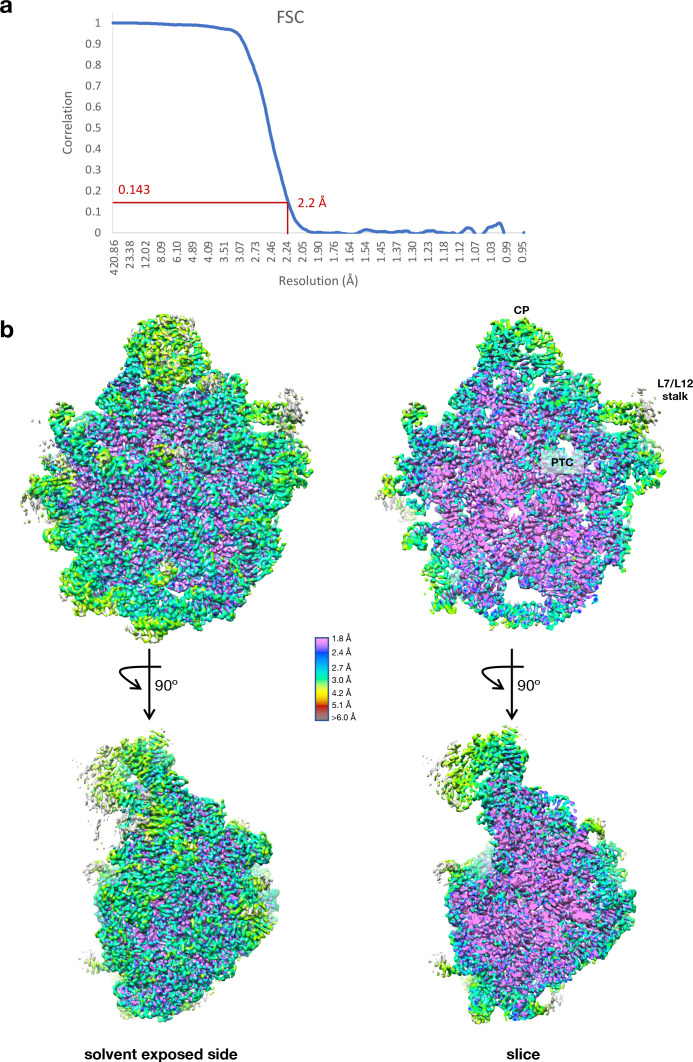
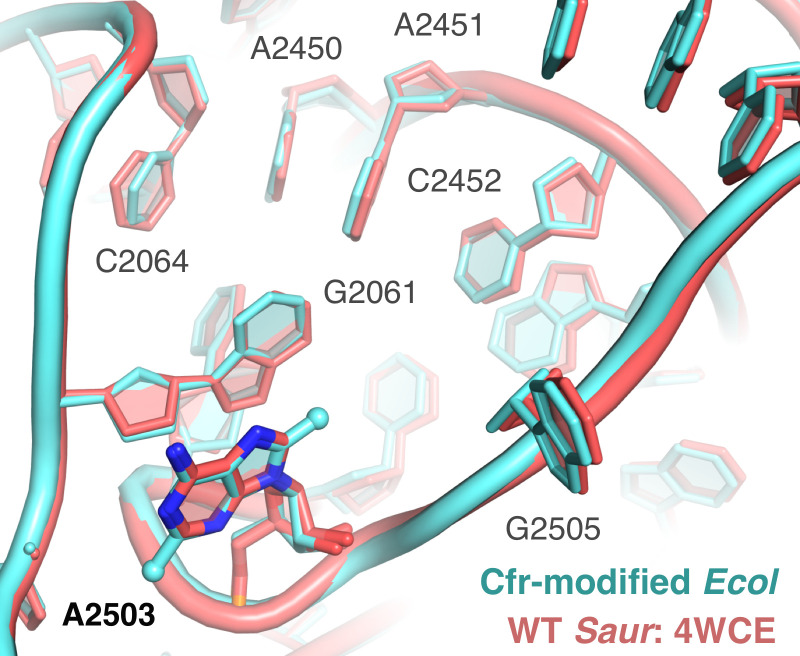
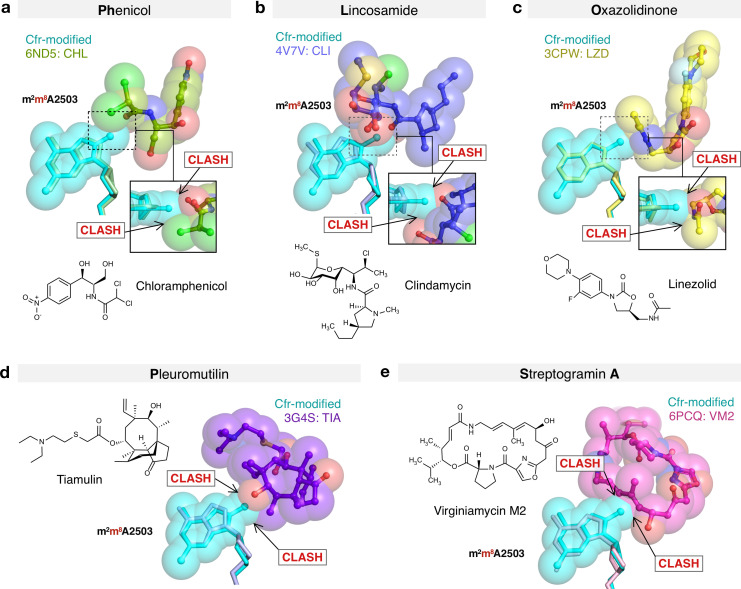
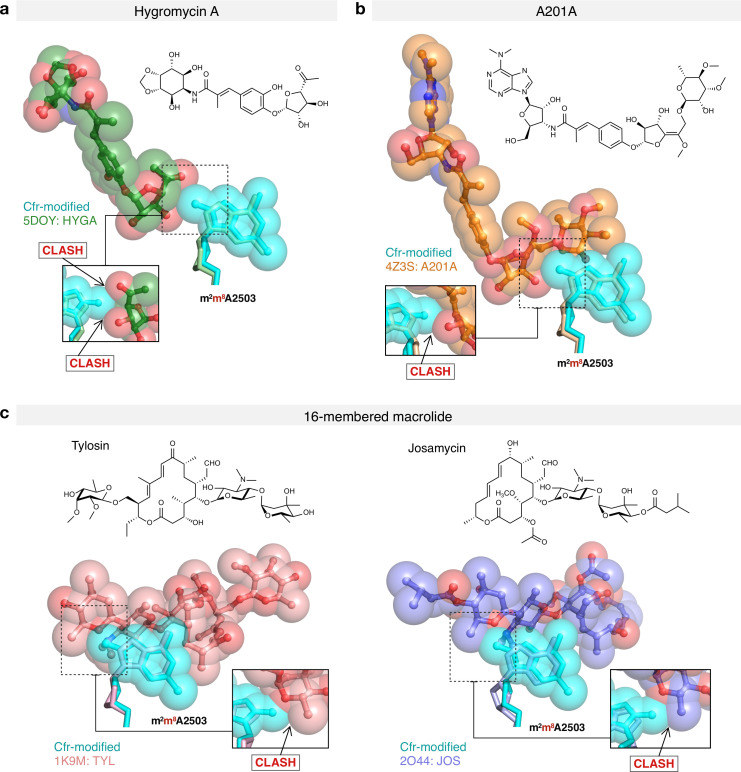
Figure 5. Near-stoichiometric ribosome methylation by CfrV7 enables structural understanding of Cfr-mediated resistance to antibiotics.
(a) Cfr-modified 50S ribosomal subunit highlighting adenosine 2503 (A2503) within 23S rRNA and the binding site of PTC-targeting antibiotics. Cfr methylates A2503 at the C8 carbon to produce m2m8A2503. (b) Cryo-EM density maps of adenosine 2503 in 23S rRNA contoured to 3σ. Cfr-modified (m2m8A2503) in cyan. Wild-type (m2A2503) in orange; PDB 6PJ6. (c) Close-up view of 23S rRNA nucleotides in the 50S ribosomal subunit. Cfr-modified Escherichia coli ribosome in cyan. Wild-type E. coli ribosome in orange; PDB 6PJ6. (d) Structural overlay of Cfr-modified E. coli ribosome (cyan) and Haloarcula marismortui 50S ribosome in complex with pleuromutilin antibiotic tiamulin (purple, PDB 3G4S) highlighting steric clashes between m8A2503 and the antibiotic. EM, electron microscopy.