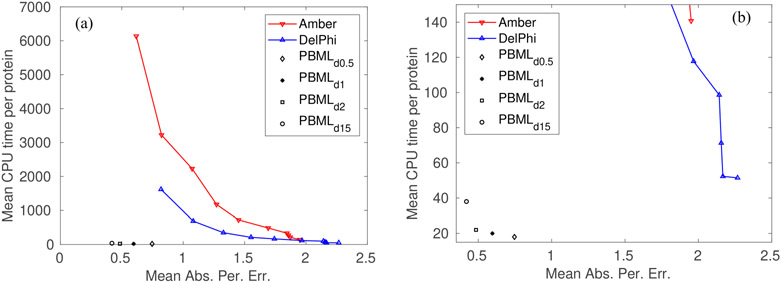
FIG. 1.
Comparison of the mean CPU time (in unit of s) per protein and the mean absolute percentage errors of Amber, DelPhi and machine learning predictions of the electrostatic solvation free energies using the test set of 195 proteins. (a) Results of Amber and DelPhi were obtained at ten different mesh sizes from 0.2 Åto 1.1 Å; Results of PBML were obtained at four MSMS densities (number of vertices per Å2) at 15, 2, 1, and 0.5. (b) A zoom-in plot of the left plot for small CPU time.