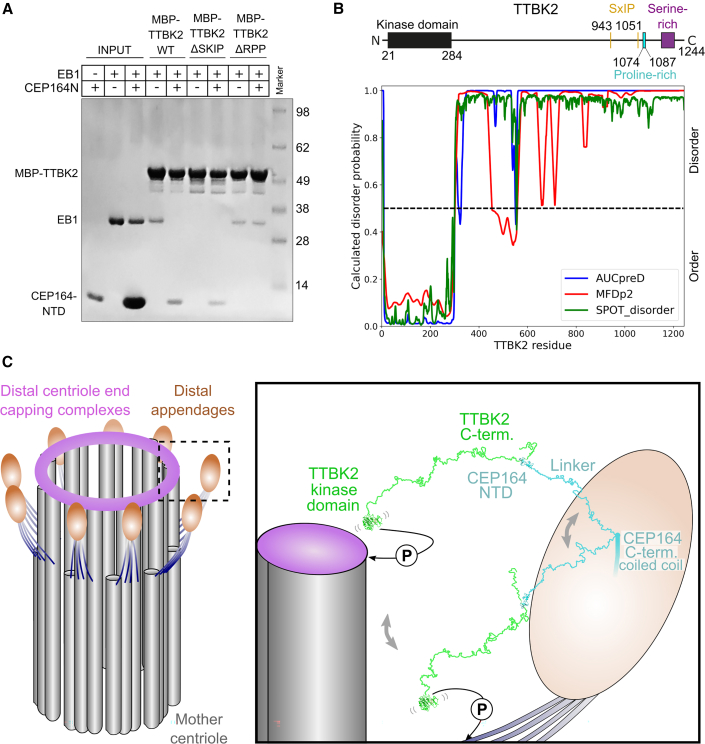
Figure 4.
CEP164-NTD binding to TTBK2 inhibits the TTBK2-EB1 interaction
(A) CEP1641−109 and EB1 compete for binding to MBP-TTBK21033−1087. Coomassie-stained SDS-PAGE gel showing pull-down experiments with purified MBP-TTBK21033−1087 (WT, EB1-binding-deficient TTBK21051−1054 SKIP to AAAA or CEP164-NTD-binding-deficient TTBK21076−1078 RPP to AAA) with EB1 in the presence of an excess of CEP1641−109.
(B) Per-residue disorder probabilities of TTBK2 as calculated by different disorder prediction algorithms. Values above 0.5 (dashed line) indicate disorder. The location of the kinase domain and selected sequence features of TTBK2 are indicated above the plot.
(C) Model of the TTBK2-CEP164 architecture at the distal appendages. Left: scheme of the mother centriole with distal appendages based on electron microscopy tomography segmentation analysis (Bowler et al., 2019). Subdistal appendages are omitted for clarity. Right: close-up view of a single distal appendage region. CEP164 is indicated in cyan, the bound TTBK2 is shown in green. The connecting linkers between CEP164-NTD and the CEP164 coiled-coil domain and between the CEP164-bound TTBK21074−1084 region and the N-terminal TTBK2 kinase domain (PDB: 6vrf) (Bao et al., 2021) are drawn in an extended and open conformation. Based on disorder predictions and the paucity of predicted secondary structure elements, we propose that these linkers are largely flexible (Figures 4B and S6B). This flexibility would allow the TTBK2-CEP164-NTD complex to sample larger areas of the distal appendage region and would enable TTBK2 to reach its phosphorylation substrates at (MPP9) or close to (CEP83) the distal centriole end. A circled “P” indicates a phosphorylation event by TTBK2.