ABSTRACT
Copper is an important component of methanotrophic physiology, as it controls the expression and activity of alternative forms of methane monooxygenase (MMO). To collect copper, some methanotrophs secrete a chalkophore- or copper-binding compound called methanobactin (MB). MB is a ribosomally synthesized posttranslationally modified polypeptide (RiPP) that, after binding copper, is collected by MbnT, a TonB-dependent transporter (TBDT). Structurally different forms of MB have been characterized, and here, we show that different forms of MB are collected by specific TBDTs. Further, we report that in the model methanotroph, Methylosinus trichosporium OB3b, expression of the TBDT required for uptake of a different MB made by Methylocystis sp. strain SB2 (MB-SB2) is induced in the presence of MB-SB2, suggesting that methanotrophs have developed specific machinery and regulatory systems to actively take up MB from other methanotrophs for copper collection. Moreover, the canonical “copper switch” in M. trichosporium OB3b that controls expression of alternative MMOs is apparent if one of the two TBDTs required for MB-OB3b and MB-SB2 uptake is knocked out, but is disrupted if both TBDTs are knocked out. These data indicate that MB uptake, including the uptake of exogenous MB, plays an important role in the copper switch in M. trichosporium OB3b and, thus, overall activity. Based on these data, we propose a revised model for the copper switch in this methanotroph that involves MB uptake.
IMPORTANCE In this study, we demonstrate that different TBDTs in the model methanotroph Methylosinus trichosporium OB3b are responsible for uptake of either endogenous MB or exogenous MB. Interestingly, the presence of exogenous MB induces expression of its specific TBDT in M. trichosporium OB3b, suggesting that this methanotroph is able to actively take up MB produced by others. This work contributes to our understanding of how microbes collect and compete for copper and also helps inform how such uptake coordinates the expression of different forms of methane monooxygenase. Such studies are likely to be very important to develop a better understanding of methanotrophic interactions via synthesis and secretion of secondary metabolites such as methanobactin and thus provide additional means whereby these microbes can be manipulated for a variety of environmental and industrial purposes.
KEYWORDS: methanotrophy, methanobactin, copper, TonB-dependent transporter, microbial interactions, chalkophore
INTRODUCTION
Methane is an important greenhouse gas with a global warming potential 28 to 34 times higher than that of carbon dioxide (1). Approximately one-third of all methane emissions are derived from natural processes, i.e., methanogenesis in pristine environments, with the rest attributed to human activities, e.g., extraction of fossil fuels, livestock farming, manure management and application, rice cultivation, and solid waste and wastewater treatment (2, 3). Annual global methane emissions have increased by over 25% in the past 40 years, largely due to human activities, making it the second most important greenhouse gas after carbon dioxide (3).
Methanotrophs, a group of microbes that use methane as their sole carbon and energy source, play an important role in eliminating methane emissions and in the global carbon cycle. Methanotrophs are phylogenetically and physiologically quite diverse, i.e., both bacteria and archaea have been found to consume methane both aerobically and anaerobically (4–10). Perhaps the best studied are the aerobic bacterial methanotrophs, i.e., members of the Proteobacteria (classes Gamma- and Alphaproteobacteria) and Verrucomicrobia phyla (11–15). These bacteria are widely distributed in diverse terrestrial, aquatic, and marine environments and play a major role in eliminating methane emissions from both natural (e.g., forest soils) as well as engineered environments (e.g., landfills) (4, 16).
The first step of methane metabolism by aerobic methanotrophs is oxidation of methane to methanol. Two forms of methane monooxygenases perform this transformation, the membrane-bound or particulate methane monooxygenase (pMMO, encoded by pmoCAB or pmo operon) and the cytoplasmic or soluble methane monooxygenase (sMMO, encoded by mmoXYBZDC or mmo operon). Interestingly, pMMO has a much higher affinity for methane than sMMO, but turns it over much more slowly (17). It may be that methanotrophs that can express both pMMO and sMMO have an advantage for growth in situ versus those methanotrophs that can only express one of the two forms (4). Interestingly, the expression and activity of the two forms of methane monooxygenase is not dependent on methane availability but, rather, copper, aka the “copper switch” (18). Under copper-limited conditions, some methanotrophs express sMMO, but as the copper/biomass ratio increases, sMMO expression is repressed, while pMMO expression increases with increasing copper (19–21). Such response is due to the fact that sMMO is a soluble di-iron monooxygenase, while pMMO has multiple metal centers; the exact nature of these is still a matter of debate, with several models proposed, although all agree that copper plays a critical role (22–26).
Some methanotrophs, including species that exhibit the copper switch (i.e., can express both sMMO and pMMO) and species that do not (i.e., only express pMMO), have been found to secrete a copper-binding compound or chalkophore termed methanobactin (MB) for copper uptake. MBs have a high affinity for copper ions and can also extract copper bound to other organic molecules as well as minerals (27–31). MBs are small (<1,300 Da) ribosomally synthesized posttranslationally modified polypeptides (RiPPs) that are characterized by two heterocyclic rings that bind copper via an N2S2 ligand set (4, 28, 32–35). MB can be divided into two general groups, groups I and II. Group I MBs have one oxazolone ring and either an oxazolone or pyrazinedione group as the second ring and form a dicyclic pyramidal-like shape after binding copper, e.g., MB from Methylosinus trichosporium OB3b (MB-OB3b) (32, 33, 36). Group II MBs have one oxazolone group with the second ring either an imidazolone or pyrazinedione ring, and they adopt a hairpin-like shape after binding copper, e.g., MB from Methylocystis sp. strain SB2 (MB-SB2) (27, 32). Recent phylogenetic analyses suggest that the two general groups can be further divided into several subgroups, i.e., groups IA, IB, IIA, IIB, and IIC (37).
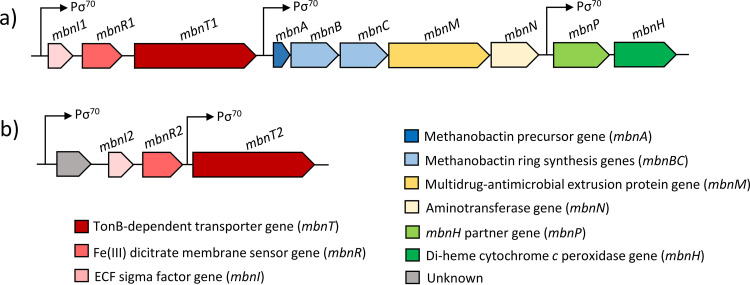
The gene encoding the polypeptide precursor of MB, mbnA, has been identified in a number of methanotrophs (27, 32, 34, 38). mbnA is part of the mbn gene cluster that includes genes involved in maturation of the MB polypeptide precursor and transport (Fig. 1a) (34, 37, 39, 40). Like sMMO and pMMO, expression of mbn genes is copper regulated, i.e., expression is greatest when copper is not provided and decreases when copper is added, i.e., expression of these genes is coregulated with sMMO and reciprocally regulated with pMMO (34, 38, 41–43). Based on various transcriptional studies, a variety of regulatory models coordinating expression of MMOs and MB have been postulated (32, 34, 42, 44).
FIG 1.
Methanobactin (a) and mbnT2 gene (b) clusters of M. trichosporium OB3b. σ70 promoter (Pσ70) regions are indicated in the gene clusters. The σ70 promoters were predicted using BPROM (73).
Also important to the discussion here is that after MB is secreted into the extracellular environment, it is subsequently taken up by a TonB-dependent transporter (TBDT) encoded by mbnT that is typically found upstream of mbnA (Fig. 1a) (37, 45, 46). Interestingly, it has been found that methanotrophs capable of making MB can also take up copper bound to MBs produced by other methanotrophs, e.g., M. trichosporium OB3b can take up copper bound to MB-SB2, and such collection regulates the expression of sMMO and pMMO (47). These data suggest that either the mechanism for MB uptake is general, i.e., MbnT associated with the MB gene cluster of M. trichosporium OB3b can take up MB-SB2, or this methanotroph can produce other TBDTs that enable it to collect MB made by other methanotrophs.
More detailed knowledge of the mechanism of copper uptake, particularly via MB in methanotrophs, is important given how copper affects methanotrophic activity. Such data would particularly help refine our understanding of how expression of key enzymes in methanotrophs are regulated and provide insight into the mechanism(s) of methanotrophic interactions, i.e., competition for copper. Such information would enhance our ability to manipulate these microbes for valorization of methane, better predict their activity in situ, and enable more advanced earth systems models of greenhouse gas emissions. Here, we investigated the function of multiple TBDTs in M. trichosporium OB3b in MB uptake and the involvement of these TBDTs in the copper switch.
RESULTS
Bioinformatic and transcriptional analyses of select TBDTs in M. trichosporium OB3b.
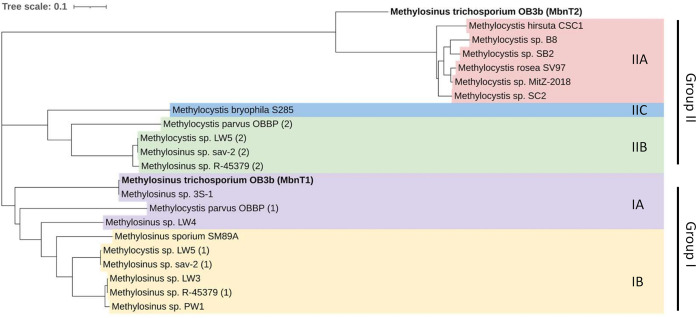
The genome of M. trichosporium OB3b encodes 30 TBDTs (Table S1 in the supplemental material), including the TBDT responsible for uptake of MB-OB3b (here labeled MbnT1). One of the other TBDTs, designated here as MbnT2, with gene locus tag ADVE02_v2_10210, has high identity (57.36%) and similarity (E value, 0.0) with MbnT from Methylocystis sp. strain SB2 (Table S1). Further, immediately upstream of mbnT2 are two other genes commonly associated with mbnTs, i.e., mbnI and mbnR, encoding an extracytoplasmic function sigma factor and a putative membrane sensor, respectively (Fig. 1b). Phylogenetic analysis of known MbnTs shows MbnT2 was related most closely to the MbnTs responsible for uptake of group IIA MBs (e.g., MB-SB2) (Fig. 2). This suggests that MbnT2 may enable M. trichosporium OB3b to take up MB-SB2, a phenomenon observed previously in M. trichosporium OB3b, but the means by which this occurs are not explained (47).
FIG 2.
Phylogenetic analysis of the amino acid sequence of putative MbnTs for methanobactin uptake. MbnTs from different groups of methanobactin are indicated with different colors. MUSCLE sequence alignment and the phylogenetic tree construction was processed using Geneious software with default settings. Further polishing of the phylogenetic tree was performed on the Interactive Tree of Life web browser (http://itol.embl.de/). The scale bar represents number of amino acid changes per site.
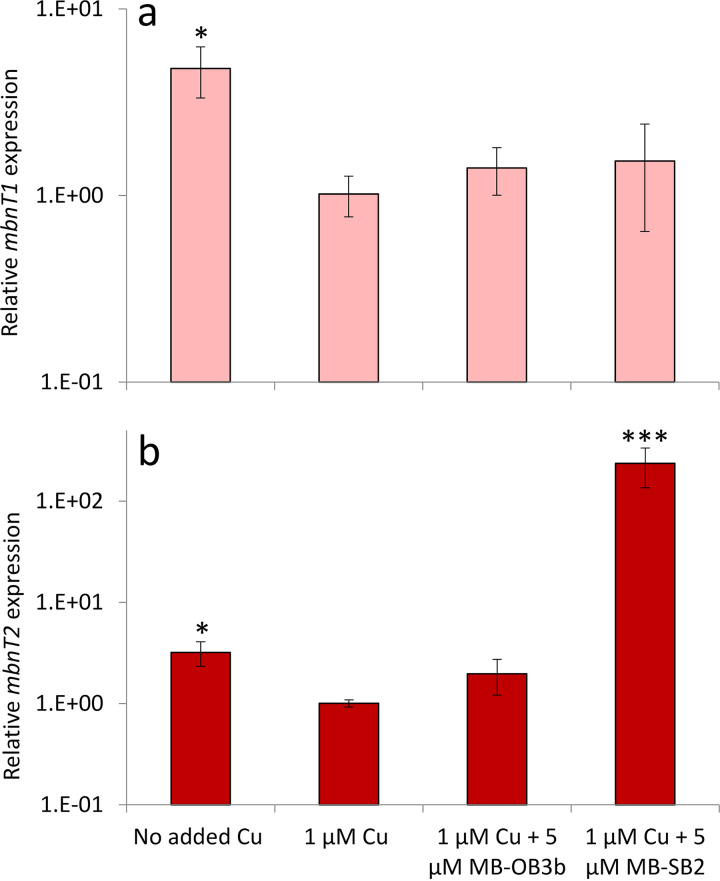
To explore the function of MbnT2 in MB-SB2 uptake, we first measured the expression of mbnT1 and mbnT2 in M. trichosporium OB3b wild type grown with different amounts of copper and MBs (Fig. 3). mbnT1 expression was approximately 5-fold higher when no copper was added (only background levels of copper, ∼30 nM) versus the addition of 1 μM copper (0.01 < P < 0.05). Further, the addition of 5 μM of either form of MB in the presence of 1 μM copper did not significantly affect mbnT1 expression compared to that observed in the presence of 1 μM copper only (Fig. 3a). Like mbnT1, mbnT2 was also upregulated when no copper was added versus in the presence of 1 μM added copper (∼3-fold; 0.01 < P < 0.05). As was found for mbnT1, the addition of 5 μM MB-OB3b in the presence of 1 μM copper did not affect mbnT2 expression. When M. trichosporium OB3b was grown with 1 μM copper plus 5 μM MB-SB2, however, mbnT2 was significantly upregulated (∼380-fold; P < 0.001) compared to the presence of 1 μM copper only (Fig. 3b).
FIG 3.
RT-qPCR analysis of the relative expression of mbnT1 (a) and mbnT2 (b) in M. trichosporium OB3b wild type grown in the absence of added copper, with 1 μM added copper, and with 1 μM added copper plus either 5 μM MB-OB3b or MB-SB2. Error bars indicate standard deviations from triplicate biological cultures. Student’s t test was performed to detect significant differences between expression in the presence of 1 μM added copper versus other growth conditions, *, 0.01 < P < 0.05; **, 0.001 < P < 0.01; ***, P < 0.001.
Deletion of TonB-dependent uptake systems in M. trichosporium OB3b.
To verify the putative function of MbnT2 in MB-SB2 uptake by M. trichosporium OB3b, a markerless deletion of mbnT2 was constructed (ΔmbnT2 mutant). mbnT2 was also deleted in the previously constructed M. trichosporium mbnT1::Gmr mutant (46) to create a double mutant defective in both MbnT1 and MbnT2 (mbnT1::Gmr ΔmbnT2 mutant). The absence of mbnT2 and the disruption of mbnT1 was confirmed by PCR (Fig. S1) and sequencing (data not shown).
Characterization of copper uptake and expression of methane monooxygenases and methanobactin production in M. trichosporium OB3b wild-type and ΔmbnT2 strains.
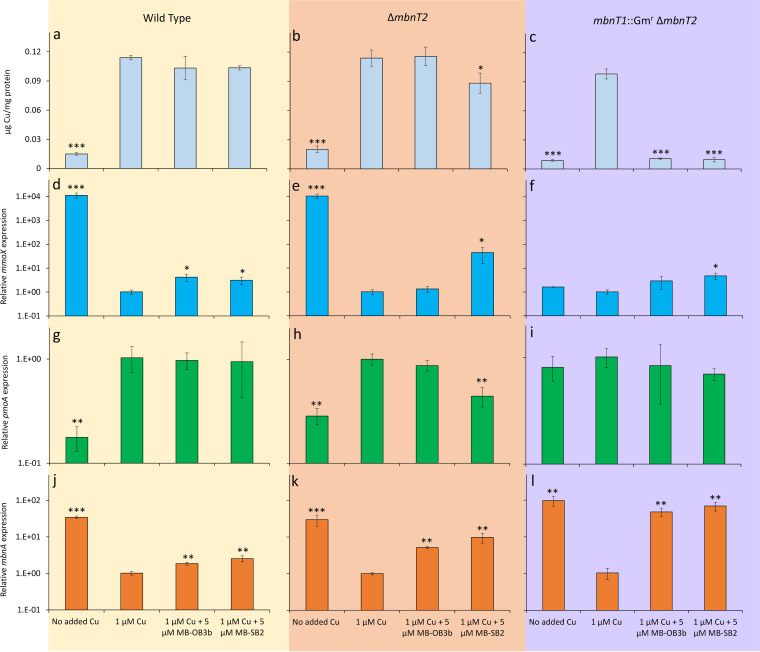
Both M. trichosporium OB3b wild-type and the ΔmbnT2 mutant strains had significantly greater copper associated with biomass when grown with 1 μM versus no added copper (Fig. 4a). There was no significant difference in the amount of copper associated with biomass for either strain when grown with 1 μM copper with 5 μM MB-OB3b versus 1 μM copper alone (Fig. 4a and b). Biomass-associated copper was also not significantly different for M. trichosporium OB3b wild type grown with 1 μM copper plus 5 μM MB-SB2 versus 1 μM copper alone, but did decrease slightly (∼20%; 0.01 < P < 0.05) in the M. trichosporium ΔmbnT2 mutant.
FIG 4.
Copper associated with biomass (a to c) and RT-qPCR analysis of the relative expression of mmoX (d to f) pmoA (g to i), and mbnA (j to l) in M. trichosporium OB3b wild type (left panels), ΔmbnT2 mutant (middle panels), and mbnT1::Gmr ΔmbnT2 mutant (right panels) grown in the absence of added copper, with 1 μM added copper, and with 1 μM added copper plus either 5 μM MB-OB3b or MB-SB2. Error bars indicate standard deviations from triplicate biological cultures. Student’s t test was performed to detect significant differences between growth in the presence of 1 μM added copper versus other growth conditions. *, 0.01 < P < 0.05; **, 0.001 < P < 0.01; ***, P < 0.001.
sMMO expression is commonly monitored via reverse transcription-quantitative PCR (RT-qPCR) of mmoX (encoding the α-subunit of the hydroxylase component of sMMO). mmoX expression was over 4 orders of magnitude greater when no copper was added versus 1 μM copper for both wild-type and ΔmbnT2 mutant strains (P < 0.001) (Fig. 4d and e). mmoX expression slightly increased in M. trichosporium OB3b wild type in the presence of 1 μM copper and 5 μM of either form of MB (an increase of 3 to 4×; 0.01 < P < 0.05), but only increased in the presence of 1 μM copper plus 5 μM MB-SB2 for the ΔmbnT2 mutant of M. trichosporium OB3b (∼45-fold; 0.01 < P < 0.05) (Fig. 4e). The naphthalene assay showed no observable sMMO activity of the ΔmbnT2 mutant cells grown with 1 μM copper and 5 μM of either form of MB (Fig. S2).
Similarly, pMMO expression is commonly monitored via RT-qPCR of pmoA (encoding the β-subunit of pMMO). pmoA expression was ∼5.8-fold-higher when 1 μM copper was added versus no copper addition and was not significantly affected with the concomitant addition of 5 μM of either MB for M. trichosporium OB3b wild type (Fig. 4g). Similarly, expression of pmoA increased ∼4-fold (0.01 < P < 0.05) with 1 μM versus no added copper for the M. trichosporium OB3b ΔmbnT2 mutant, and such expression did not significantly change with the addition of MB-OB3b (Fig. 4h). pmoA expression, however, decreased by ∼55% (0.001 < P < 0.01) in the presence of 1 μM copper plus 5 μM MB-SB2 compared to 1 μM copper alone in the M. trichosporium OB3b ΔmbnT2 mutant (Fig. 4g and h).
mbnA expression increased ∼35-fold (P < 0.001) when no copper was added versus in the presence of 1 μM added copper for M. trichosporium OB3b wild type (Fig. 4j). The addition of 5 μM of either form of MB in the presence of copper also increased mbnA expression in M. trichosporium OB3b wild type compared to the addition of copper alone (1.8 to 2.5×; 0.001 < P < 0.01). Expression of mbnA in M. trichosporium OB3b ΔmbnT2 similarly was ∼30× higher when no copper was added versus in the presence of 1 μM added copper (P < 0.001; Fig. 4k). Further, mbnA expression increased in M. trichosporium OB3b ΔmbnT2 in the presence of 1 μM copper and 5 μM of either form of MB compared to copper alone (0.001 < P < 0.01), but expression increased more in the presence of MB-SB2 (∼10×) versus MB-OB3b (∼5×).
Characterization of copper uptake and expression of methane monooxygenases and methanobactin production in M. trichosporium OB3b mbnT1::Gmr ΔmbnT2.
To further delineate the mechanism(s) underlying MB uptake by M. trichosporium OB3b, we constructed and characterized the double mutant of mbnT1::Gmr ΔmbnT2. Copper associated with biomass when no additional copper was provided was an order of magnitude less than in the presence of 1 μM copper for this mutant (∼12-fold less, P < 0.001). Copper uptake was similarly reduced by an order of magnitude when 5 μM of either form of MB was provided in addition to 1 μM copper compared to copper alone (P < 0.001) (Fig. 4c). Further, the addition of 1 μM copper had little effect on either mmoX or pmoA expression in this double mutant compared to when no copper was added (Fig. 4f and i). Expression of mmoX increased ∼3× when 5 μM MB-OB3b was added in conjunction with 1 μM copper, but this was not significantly different from when only copper was added (P > 0.05). mmoX expression in the mbnT1::Gmr ΔmbnT2 mutant increased more when 5 μM MB-SB2 was added in conjunction with copper and was significantly different from when only copper was added (∼5×; 0.01 < P < 0.05). pmoA expression was not affected by the addition of either form of MB in the presence of 1 μM copper. Collectively, these data indicate that the copper switch was disrupted in this strain, i.e., mmo and pmo gene expression did not respond to the addition/nonaddition of copper. Finally, expression of mbnA was 2 orders of magnitude greater (0.001 < P < 0.01) when no copper was added than in the presence of 1 μM added copper in this double mutant (Fig. 4l). Cultivation of this mutant in the presence of 1 μM copper plus 5 μM of either form of MB increased mbnA expression by ∼50 to 70× compared to the presence of copper alone (0.001 < P < 0.01) (Fig. 4l).
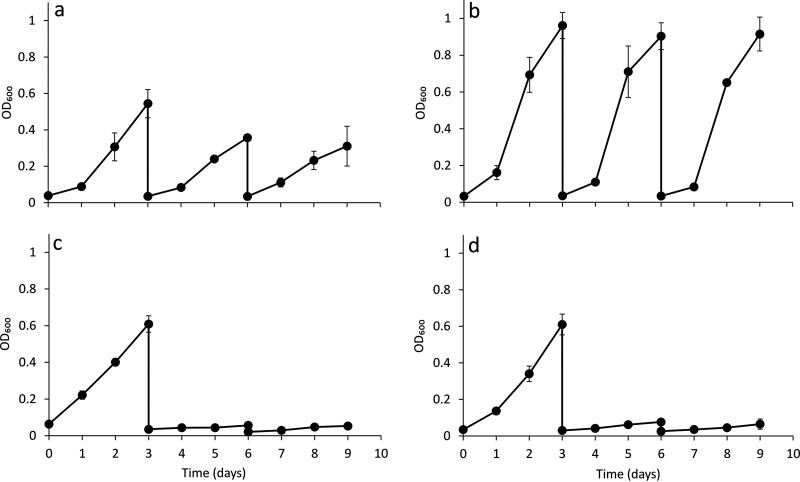
Further growth experiments confirmed that of mbnT1::Gmr ΔmbnT2 mutant of M. trichosporium OB3b was likely unable to carefully regulate expression of the alternative forms of MMO. That is, growth of this double mutant in methane was observed if transferred multiple times without extra copper addition or in the presence of 1 μM added copper (Fig. 5a and b). sMMO activity, however, was not apparent via the naphthalene assay when copper was not added when characterized at the end of the third cycle (Fig. S3). If this mutant was grown in the presence of 1 μM copper and 5 μM of either form of MB, however, growth stopped after a second sequential transfer (Fig. 5c and d). These data suggest that this mutant is unable to collect copper in the presence of either form of MB but does have some mechanism(s) to scavenge trace amounts of copper in their absence. Growth, however, was inhibited when no copper was added compared to growth in the presence of 1 μM copper, i.e., the maximum cell density of the mbnT1::Gmr ΔmbnT2 mutant of M. trichosporium OB3b grown with only background levels of copper (∼30 nM) was ∼30% of the biomass when grown with 1 μM copper (Fig. 5a and b). It also appears, based on RT-qPCR and sMMO activity assays, that this mutant is unable to effectively utilize sMMO for methane oxidation, especially when copper is completely unavailable (i.e., through exogenous provision of MB) and instead constitutively expresses (and solely relies on) pMMO activity for growth.
FIG 5.
Growth of M. trichosporium mbnT1::Gmr ΔmbnT2 mutant in the absence of added copper (a), with 1 μM added copper (b), with 1 μM added copper plus 5 μM MB-OB3b (c), or with 1 μM added copper plus 5 μM MB-SB2 (d). Error bars indicate standard deviations from triplicate biological cultures.
These results indicate that in the M. trichosporium OB3b mbnT1::Gmr ΔmbnT2 mutant, the copper switch was disrupted. To investigate this further, both M. trichosporium OB3b wild-type and mbnT1::Gmr ΔmbnT2 mutant strains were grown with methanol (0.5%) as the growth substrate and various amounts of copper (either no addition or 1 μM). As observed earlier (18), the copper switch was clearly apparent in M. trichosporium OB3b wild type when grown in the presence of methanol with and without extra addition of 1 μM copper (Fig. S4a, c, and e). Specifically, mmoX expression and sMMO activity clearly responded to the addition/nonaddition of copper in the wild-type strain when grown with methanol (Fig. S4a and e). Conversely, as was found for methane-grown cultures, expression of pmoA in M. trichosporium OB3b wild type increased by approximately 8-fold when 1 μM copper was added versus no copper (0 μM) addition. When the M. trichosporium OB3b mbnT1::Gmr ΔmbnT2 mutant strain was grown on methanol and various amounts of copper, again, as found for methane-grown cultures, mmoX expression was invariant with respect to copper, and the naphthalene assay did not indicate any sMMO activity under any condition (Fig. S4b and f). pmoA expression for methanol-grown cultures of M. trichosporium OB3b mbnT1::Gmr ΔmbnT2, however, did increase ∼3× when grown in the presence of 1 μM copper versus no added copper (Fig. S4d). Such a difference was significant (P = 0.013) but less than that observed in M. trichosporium OB3b wild type grown on methanol (where pmoA expression increased ∼8× in the presence of 1 μM copper versus no added copper; P = 0.0007) (Fig. S4c).
DISCUSSION
MBs have been traditionally categorized into one of two general groups based on general chemical composition and structure. Recent phylogenetic analyses suggest that these groups can be further subdivided into at least five smaller groups, i.e., groups IA, IB, IIA, IIB, and IIC, although what, if any, structural differences that may exist between these subgroups is unclear, as MBs to date have only been characterized from groups IA and IIA (37). Our phylogenetic analyses of TBDTs involved in MB uptake (MbnTs), however, suggest that these also can generally be divided into groups that correspond to the proposed MB subgrouping system, suggesting that these MB subgroups may indeed have different structures (Fig. 2). That is, the general correspondence of grouping of the genes involved in MB biosynthesis with MB uptake suggests that MbnTs preferentially bind and import specific forms of MB due to different structures these MBs make after binding copper. Such a phenomenon is quite possible, as it has been shown that different siderophores are taken up preferentially by different TBDTs (48, 49).
Previous studies indeed demonstrated MbnT1 encoded by the mbn gene cluster of M. trichosporium OB3b is the only TBDT responsible for MB-OB3b uptake (45, 46). Here, we show, using various mutants, that MbnT2 preferentially binds and transports MB-SB2 (a group IIA MB), but we cannot exclude, at this time, that MB-SB2 may also be collected to some degree by M. trichosporium OB3b via MbnT1. That is, although copper uptake was slightly yet significantly reduced (∼20%) in the M. trichosporium OB3b ΔmbnT2 mutant in the presence of MB-SB2, it was still apparent. Such a hypothesis is supported by the finding that copper uptake was effectively abolished in the double mutant of M. trichosporium OB3b mbnT1::Gmr ΔmbnT2 in the presence of either MB-OB3b or MB-SB2. Such a conclusion, however enticing, must be treated with some caution, as both the M. trichosporium OB3b ΔmbnT2 and M. trichosporium OB3b mbnT1::Gmr ΔmbnT2 mutants are still able to produce MB-OB3b, as expression of the biosynthetic genes for MB production is under the control of a separate promoter from mbnT1 (Fig. 1). An alternative explanation may be that MB-OB3b can extract copper to some degree from MB-SB2, thus allowing for some copper uptake by M. trichosporium OB3b ΔmbnT2 via mbnT1 in the presence of copper and MB-SB2, but it was reduced compared to the presence of copper only. We cannot reject either hypothesis at this time, and more effort is clearly needed to address this point.
As found before, expression of MB biosynthesis and uptake genes (i.e., mbnA, mbnT1, and mbnT2) were regulated by the availability of copper, with expression decreasing when copper was added versus no added copper (41). Such a finding is logical, for, as copper availability increases, the need to express high-affinity systems such as MB to collect copper is relaxed. Interestingly, here, we find that mbnT1 expression in M. trichosporium OB3b was not significantly affected by the presence of either MB-OB3b or MB-SB2, but mbnT2 expression dramatically increased in the presence of MB-SB2 and copper. This suggests that M. trichosporium OB3b has an active mechanism to sense and collect MBs made by other methanotrophs. Such a possibility is not unprecedented, as many microorganisms have been found to take up siderophores to satisfy iron requirements, and the systems required for such uptake are often induced by the presence of heterologous siderophores (50–57).
In addition to participating in uptake of copper in the presence of MBs, it appears that MbnT1 and MbnT2 collectively play a very important role in the canonical copper switch in M. trichosporium OB3b. That is, it was earlier shown that mutants of M. trichosporium OB3b, either defective in producing MB or in its uptake (i.e., disruption of mbnA or mbnT, respectively), still differentially expressed sMMO and pMMO, as copper availability was varied (34, 46). If mbnT2 was deleted, again, the copper switch was apparent when no copper was added versus in the presence of 1 μM added copper (Fig. 4). When both mbnT1 and mbnT2 were knocked out, however, the copper switch was disrupted in that mmoX expression in this mutant did not respond to copper, and activity of sMMO was never apparent, even when copper was not added for both methane and methanol-grown cultures of the M. trichosporium OB3b mbnT1::Gmr ΔmbnT2 mutant (Fig. 4; Fig. S3 and S4). Further, pmoA expression also did not respond to the addition of 1 μM copper in this mutant when grown on methane (Fig. 4), but there was some response for methanol-grown cultures when 1 μM copper was added. However, such an increase was much less than that observed in M. trichosporium OB3b wild type.
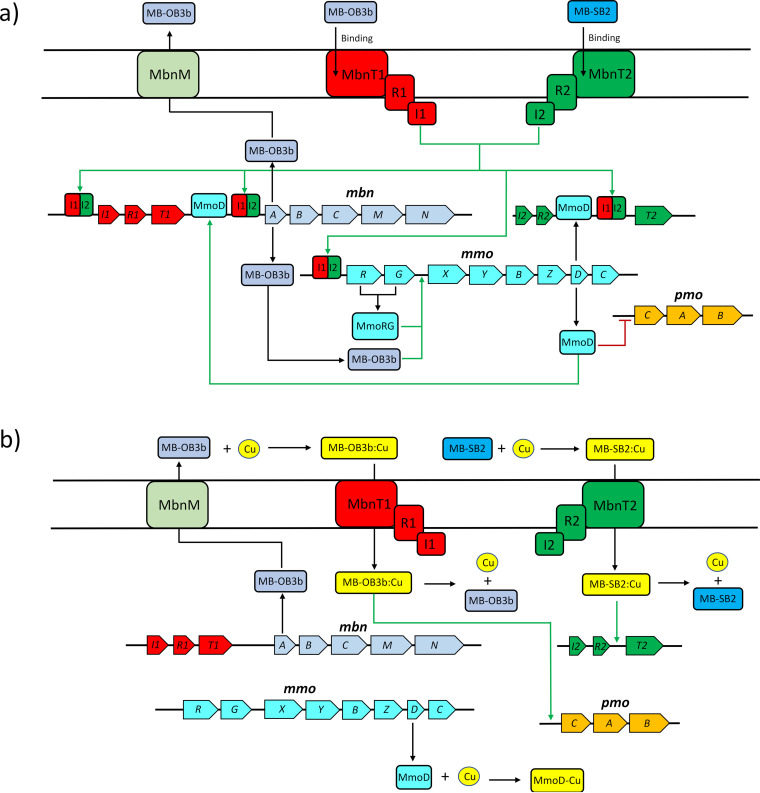
It should be noted that multiple regulatory models for the copper switch have been proposed. One presumes that the switch is based on a FecIRA-like signal cascade similar to that found in siderophore synthesis. That is, as shown in Fig. 1, both mbnT1 and mbnT2 are part of a FecIRA-like gene cluster where immediately upstream of these mbnTs are genes encoding a putative membrane sensor (mbnR) and an extracytoplasmic sigma factor (mbnI). In many siderophore production systems, after an outer membrane transporter such as TBDT binds a ferrisiderophore, a signal cascade is observed where the outer membrane transporter transmits a signal to the membrane sensor that activates the extracytoplasmic sigma factor, subsequently inducing further siderophore production (and in some cases, other unrelated genes) (58, 59). Given the strong similarity in gene organization between MB and siderophore uptake systems, it has been proposed that an analogous regulatory system exists in methanotrophs that express MB (42). Such a model, although certainly appropriate for iron-regulated gene expression in some microbes, is not supported by previous data that showed disruption of mbnT1 did not affect the copper switch (46). Rather, the original model proposed for the copper switch is that it is based on MmoD, a polypeptide encoded within the mmo operon of those methanotrophs that exhibit the copper switch and that MB (and any signal cascade associated with its uptake) serves to amplify the magnitude of the switch (34).
The data presented here provide the basis of a third model that may reconcile these conflicting proposals for the copper switch. That is, when MB uptake was prevented through the knockout of both mbnT1 and mbnT2, the copper switch was clearly disrupted. It thus appears that in M. trichosporium OB3b, the copper switch has multiple layers of control, i.e., M. trichosporium OB3b has redundant mechanisms to collect copper via MB and TBDT (MbnT1 and MbnT2), and only if both are disrupted is the copper switch no longer operative. It thus appears that one TBDT for MB uptake, along with associated regulatory genes (i.e., mbnI and mbnR), is sufficient to maintain the copper switch in conjunction with MmoD. Given these new data, we present an updated model for the copper switch in Fig. 6. Here, it is proposed that in the absence of significant amounts of copper, MmoD represses pmo gene expression, while the binding of copper-free MB to MbnT activates MbnI that upregulates expression of known regulatory elements of the mmo operon (MmoR and MmoG). This is possible, as it has been found that iron-free siderophores can be bound to specific TBDTs, inducing gene expression (55, 60). MmoR and MmoG then, in conjunction with MB, may increase expression of genes encoding various sMMO polypeptides as well as MmoD. Further, MbnI, along with MmoD, is proposed to upregulate expression of mbn genes. When copper is present, copper-MB complexes can be taken up that inhibit the ability of signal transduction from MbnT through MbnR to MbnI, and thus, expression of mbn and mmo genes is reduced. After copper-MB uptake, copper is released and associated with MmoD that then serves to prevent repression of the pmo operon. In addition, based on earlier findings, copper-MB complexes may also serve to enhance expression of the pmo operon, but this is not critical for such expression (34).
FIG 6.
Proposed regulatory scheme of mmo, pmo, and mbn operons in M. trichosporium OB3b grown in the absence of added copper (a) or with copper plus MB-OB3b or copper plus MB-SB2 (b). Red lines and green arrows indicate repression or promotion of the gene transcription.
Such a model is intriguing, as it indicates that methanotrophs have not only evolved high-affinity means to collect copper (i.e., MB), but at least one (M. trichosporium OB3b) has developed a unique regulatory system to recognize and actively take up MB produced by another methanotroph by upregulating the system required for its uptake. Upregulation of mbnT1 was not observed when additional MB-OB3b was provided, suggesting that there are some critical, as-yet-unknown regulatory differences between MbnT1/T2, MbnR1/R2, and/or MbnI1/I2 to enable M. trichosporium OB3b to selectively upregulate mbnT2 in the presence of MB-SB2 but not mbnT1 in the presence of MB-OB3b. It is thus still unclear how M. trichosporium OB3b is able to differentially respond to different forms of MB. An additional issue that should be considered is that although our data indicate that specific TBDTs target different forms of MB to take up, we cannot exclude the possibility that MbnIR associated with one MbnT cannot form an effective complex with the other to enable a signal cascade to be created. Such a possibility is intriguing, as it would again indicate that methanotrophs that exhibit the copper switch have multiple layers of control to ensure appropriate expression of MMOs dependent on copper availability.
In conclusion, we show that MB is important for copper collection in methanotrophs, and some individuals are not only able to produce and collect their own form of MB but also have evolved specific machinery and regulatory systems to actively take up MB from others. Such character may have two purposes, i.e., it would enable the recipient to (i) capture copper bound to various forms of MB that are usually unavailable for many microorganisms, including methanotrophs and denitrifiers (61, 62); and (ii) improve its competitiveness by placing other MB-producing methanotrophs at a disadvantage. These studies also expand our understanding of how methanotrophs interact by elucidating that different forms of MB require different uptake systems and that at least one methanotroph utilizes multiple MB uptake systems to create substantial redundancy in the copper switch that plays a critical role in its metabolism. Such information has significant potential for enhancing future efforts aimed at manipulating methanotrophs for a number of lucrative and important purposes.
MATERIALS AND METHODS
Methanobactin isolation.
Methanobactin (MB) from M. trichosporium OB3b and Methylocystis sp. strain SB2 were isolated from their spent media as previously described by Bandow et al. (63). The purity of the isolated methanobactins was determined by high-performance liquid chromatography (HPLC) as described earlier (64).
Growth conditions.
Methylosinus trichosporium OB3b and constructed mutants (Table 1) were grown in nitrate mineral salt (NMS) medium (6) with or without 1 μM CuCl2. For NMS medium without added copper, the background copper concentration was found to be 30 ± 10 nM. Methane and air were added at a methane-to-air ratio of 1:2. For methanol grown cultures, 0.5% (vol/vol) methanol was added to the NMS medium as described before (18). Cultures were incubated in the dark at 30°C. Liquid cultures were grown in 250-ml sidearm Erlenmeyer flasks with 20 to 30 ml NMS medium with shaking at 200 rpm. Methanobactin from M. trichosporium OB3b or Methylocystis sp. strain SB2 were filter sterilized and added to NMS medium at a final concentration of 5 μM as described earlier (65). Solid NMS medium was supplemented with 1.2% agar. Growth was monitored by measuring the optical density at 600 nm (OD600) with a Genesys 20 visible spectrophotometer (Spectronic Unicam, Waltham, MA). Triplicate biological cultures were harvested at mid- to late-exponential phase for OD600 measurement, transcriptional analysis of specific gene expression, and metal distribution. Escherichia coli was grown in Luria-Bertani broth (LB) at 37°C with or without supplementation of 25 μg/ml kanamycin.
TABLE 1.
Bacterial strains used in this study
| Strain | Description and/or genotype | Reference or source |
|---|---|---|
| Methylocystis sp. strain SB2 | Wild type | 74 |
| M. trichosporium OB3b | Wild type | |
| M. trichosporium mbnT1::Gmr | mbnT1 partially exchanged with gentamicin resistance gene | 46 |
| M. trichosporium ΔmbnT2 | mbnT2 deleted | This work |
| M. trichosporium mbnT1::Gmr ΔmbnT2 | mbnT1 partially exchanged with gentamicin resistance gene, and mbnT2 deleted | This work |
| E. coli TOP10 | Strain used for plasmid construction and cloning. F-mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara leu) 7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| E. coli S17-1 | Conjugative donor. recA1 thi pro hsdR-RP4-2Tc::Mu Km::Tn7 | 67 |
Construction of M. trichosporium OB3b mutants.
mbnT2 was knocked out using a previously described protocol (66) with modifications. Briefly, upstream and downstream regions of mbnT2 (arms A and B, respectively) were PCR amplified using the arm primers listed in Table 2. Arms A and B were digested with the restriction enzymes and ligated together to form armAB, which was subsequently inserted into pK18mobsacB mobilizable suicide vector (Fig. S5) (67). The pK18mobsacB vector with armAB was transferred to E. coli TOP10 (Invitrogen, Carlsbad, CA). Plasmid was extracted from transformed E. coli Top10 using the plasmid minikit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. The extracted plasmid was then transferred to Escherichia coli S17-1 (68). Conjugation of E. coli S17-1 carrying the constructed vector with M. trichosporium OB3b was performed as described by Martin and Murrell (69). Transconjugants were grown on NMS plates supplemented with 25 μg/ml kanamycin and 10 μg/ml nalidixic acid. One kanamycin-resistant transconjugant colony (generated after 10 days incubation) was transferred to an NMS plate with kanamycin (25 μg/ml) and incubated for 7 days and subsequently transferred to an NMS plate with 2.5% sucrose (mass/vol). Sucrose-resistant colonies were generated after 10 days incubation and were screened for mutant with deletion of mbnT2 by colony PCR using the checking primers (Table 2). The successful mbnT2 deletion mutant was further confirmed by PCR with DNA extracted from the mutant using the DNeasy PowerSoil Pro kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. The procedure for construction and confirmation of the mbnT1::Gmr ΔmbnT2 double mutant was the same as described above, except using mbnT1::Gmr (46) mutant strain instead of the wild-type M. trichosporium OB3b for the conjugation step.
TABLE 2.
Primers used in this study
| Targeted gene | Primers (sequence [5′–3′])a | Application | Reference or source |
|---|---|---|---|
| mbnT1 | mbnT1-checking_F (CCGATCGAACCTGGCTCTAT), mbnT1-checking_R (ATTGTAAATCGTGACGGCGG) | PCR | 46 |
| mbnT1 | qmbnT1_F (TATGCGCCGGTCTATGGTTC), qmbnT1_R (GCCGAGATCATGTCCTGGAG) | RT-qPCR | This work |
| mbnT2 | mbnT2_F (CTGAAGACCGTGAATCCGCT), mbnT2_R (GTCCATTGGCCTGTGTGAGA) | PCR | This work |
| mbnT2 | mbnT2-armA_F, (ATTTTT gaattc ACATGAGCAACGGCCAGAAA)b, mbnT2-armA_R (ATTTTT cactttgtg GAACGCGGAACCTCCTTCA)b | PCR | This work |
| mbnT2 | mbnT2-armB_F (ATTTTT cacaaagtg TGAATGGTTCCGCAACGAGA)b, mbnT2-armB_R (ATTTTT aagctt AGAAAAGGCCGCCTACCTTC)b | PCR | This work |
| mbnT2 | qmbnT2_F (GCAATATAGTCCCGGCGTGT), qmbnT2_R (AGAAAGGGTATGTCGTGCCG) | RT-qPCR | This work |
| pmoA | pmoA_F (TTCTGGGGCTGGACCTAYTTC), pmoA_R (CCGACAGCAGCAGGATGATG) | RT-qPCR | 75 |
| mmoX | mmoX_F (TCAACACCGATCTSAACAACG), mmoX_R (TCCAGATTCCRCCCCAATCC) | RT-qPCR | 75 |
| mbnA | mbnA-checking_F (GCGATCAAGTAGGTATAACTTGGAA), mbnA-checking_R (CAATTCCTCCCGATCTCTTTC) | PCR | 39 |
| mbnA | qmbnA_F (TGGAAACTCCCTTAGGAGGAA), qmbnA_R (CTGCACGGATAGCACGAAC) | RT-qPCR | 34 |
| 16S rRNA | Eub_341F (CCTACGGGAGGCAGCAG) Eub_534R (ATTACCGCGGCTGCTGGC) | RT-qPCR | 76 |
Y, S, and R are the IUPAC DNA codes for the C/T, C/G, and A/G nucleobases, respectively.
Lowercase letters indicate EcoRI, AdeI, or HindIII restriction site sequences included in these primers.
RNA isolation and RT-qPCR.
RNA isolation was performed with a bead-beating procedure followed by column purification using RNeasy minikit (Qiagen, Hilden, Germany) as described before (70). Genomic DNA was removed on the column with RNase-free DNase (Qiagen, Hilden, Germany) treatment. Absence of genomic DNA was confirmed by 16S rRNA gene-targeted PCR with extracted RNA samples as the templates. The purified RNA was quantified using a NanoDrop 1000 spectrophotometer (Thermo Scientific, Wilmington, DE). cDNA was synthesized from 200 ng total RNA using SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA) following the manufacturer’s instructions.
RT-qPCR was performed to determine the relative expression of the mbnT1, mbnT2, pmoA, mmoX, and mbnA in M. trichosporium OB3b and mutant strains grown with and without addition of copper and/or methanobactins. Primers for amplification of the two mbnT genes were designed using the NCBI online primer design tool (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). Primer specificity was checked with the online tool and further verified by PCR, gel electrophoresis, and sequencing. RT-qPCR was performed using the iTaq Universal SYBR green supermix (Bio-Rad, Hercules, CA) with 96-well PCR plates on a CFX Connect real-time PCR detection system (Bio-Rad, Hercules, CA). The RT-qPCR program was 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, 56°C for 30 s, and 72°C for 30 s. Melting curves were measured from 65°C to 95°C with increments of 0.5°C and 10 s at each step. Transcription of the targeted genes was determined using cDNA as the template. The transcript levels were calculated by relative quantification using the quantification cycle (2−ΔΔCq) method (71) with the 16S rRNA gene as the reference gene (72).
Metal analysis.
Cells of M. trichosporium OB3b and the mutant strains grown under different conditions were collected and acid digested as described early (46, 72). Copper associated with biomass was subsequently analyzed using an inductively coupled plasma mass spectrometer (ICP-MS; Agilent Technologies, Santa Clara, CA).
Data availability.
Materials and data generated in this study will be made available upon reasonable request to the corresponding author.
ACKNOWLEDGMENTS
This research was supported by the U.S. Department of Energy Office of Science (grant no. DE-SC0020174 to J.D.S. and A.A.D.) and the National Science Foundation (grant no. 1724744 to J.D.S.). Lawrence Livermore National Laboratory is operated by Lawrence Livermore National Security, LLC, for the U.S. Department of Energy, National Nuclear Security Administration, under contract DE-AC52-07NA27344. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We declare we have no competing interests.
Footnotes
Supplemental material is available online only.
Contributor Information
Jeremy D. Semrau, Email: jsemrau@umich.edu.
Isaac Cann, University of Illinois at Urbana-Champaign.
REFERENCES
- 1.Intergovernmental Panel on Climate Change (IPCC). 2013. Climate change 2013: the physical science basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge University Press, Cambridge, UK. [Google Scholar]
- 2.Nisbet EG, Dlugokencky EJ, Bousquet P. 2014. Methane on the rise—again. Science 343:493–495. 10.1126/science.1247828. [DOI] [PubMed] [Google Scholar]
- 3.Saunois M, Stavert AR, Poulter B, Bousquet P, Canadell JG, Jackson RB, Raymond PA, Dlugokencky EJ, Houweling S, Patra PK, Ciais P, Arora VK, Bastviken D, Bergamaschi P, Blake DR, Brailsford G, Bruhwiler L, Carlson KM, Carrol M, Castaldi S, Chandra N, Crevoisier C, Crill PM, Covey K, Curry CL, Etiope G, Frankenberg C, Gedney N, Hegglin MI, Höglund-Isaksson L, Hugelius G, Ishizawa M, Ito A, Janssens-Maenhout G, Jensen KM, Joos F, Kleinen T, Krummel PB, Langenfelds RL, Laruelle GG, Liu L, Machida T, Maksyutov S, McDonald KC, McNorton J, Miller PA, Melton JR, Morino I, Müller J, Murguia-Flores F, et al. 2020. The global methane budget 2000–2017. Earth Syst Sci Data 12:1561–1623. 10.5194/essd-12-1561-2020. [DOI] [Google Scholar]
- 4.Semrau JD, DiSpirito AA, Gu W, Yoon S. 2018. Metals and methanotrophy. Appl Environ Microbiol 84:e02289-17. 10.1128/AEM.02289-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Timmers PH, Welte CU, Koehorst JJ, Plugge CM, Jetten MS, Stams AJ. 2017. Reverse methanogenesis and respiration in methanotrophic archaea. Archaea 2017:1654237. 10.1155/2017/1654237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whittenbury R, Phillips K, Wilkinson J. 1970. Enrichment, isolation and some properties of methane-utilizing bacteria. Microbiology 61:205–218. 10.1099/00221287-61-2-205. [DOI] [PubMed] [Google Scholar]
- 7.Boetius A, Ravenschlag K, Schubert CJ, Rickert D, Widdel F, Gieseke A, Amann R, Jørgensen BB, Witte U, Pfannkuche O. 2000. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407:623–626. 10.1038/35036572. [DOI] [PubMed] [Google Scholar]
- 8.Hinrichs K-U, Hayes JM, Sylva SP, Brewer PG, DeLong EF. 1999. Methane-consuming archaebacteria in marine sediments. Nature 398:802–805. 10.1038/19751. [DOI] [PubMed] [Google Scholar]
- 9.Orphan VJ, House CH, Hinrichs K-U, McKeegan KD, DeLong EF. 2001. Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 293:484–487. 10.1126/science.1061338. [DOI] [PubMed] [Google Scholar]
- 10.Knittel K, Lösekann T, Boetius A, Kort R, Amann R. 2005. Diversity and distribution of methanotrophic archaea at cold seeps. Appl Environ Microbiol 71:467–479. 10.1128/AEM.71.1.467-479.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Op den Camp HJ, Islam T, Stott MB, Harhangi HR, Hynes A, Schouten S, Jetten MS, Birkeland NK, Pol A, Dunfield PF. 2009. Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ Microbiol Rep 1:293–306. 10.1111/j.1758-2229.2009.00022.x. [DOI] [PubMed] [Google Scholar]
- 12.Dunfield PF, Yuryev A, Senin P, Smirnova AV, Stott MB, Hou S, Ly B, Saw JH, Zhou Z, Ren Y, Wang J, Mountain BW, Crowe MA, Weatherby TM, Bodelier PLE, Liesack W, Feng L, Wang L, Alam M. 2007. Methane oxidation by an extremely acidophilic bacterium of the phylum Verrucomicrobia. Nature 450:879–882. 10.1038/nature06411. [DOI] [PubMed] [Google Scholar]
- 13.Islam T, Jensen S, Reigstad LJ, Larsen Ø, Birkeland N-K. 2008. Methane oxidation at 55°C and pH 2 by a thermoacidophilic bacterium belonging to the Verrucomicrobia phylum. Proc Natl Acad Sci USA 105:300–304. 10.1073/pnas.0704162105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pol A, Heijmans K, Harhangi HR, Tedesco D, Jetten MS, Den Camp HJO. 2007. Methanotrophy below pH 1 by a new Verrucomicrobia species. Nature 450:874–878. 10.1038/nature06222. [DOI] [PubMed] [Google Scholar]
- 15.van Teeseling MC, Pol A, Harhangi HR, van der Zwart S, Jetten MS, den Camp HJO, van Niftrik L. 2014. Expanding the verrucomicrobial methanotrophic world: description of three novel species of Methylacidimicrobium gen. nov. Appl Environ Microbiol 80:6782–6791. 10.1128/AEM.01838-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Semrau JD, DiSpirito AA, Yoon S. 2010. Methanotrophs and copper. FEMS Microbiol Rev 34:496–531. 10.1111/j.1574-6976.2010.00212.x. [DOI] [PubMed] [Google Scholar]
- 17.Lee S-W, Keeney DR, Lim D-H, Dispirito AA, Semrau JD. 2006. Mixed pollutant degradation by Methylosinus trichosporium OB3b expressing either soluble or particulate methane monooxygenase: can the tortoise beat the hare? Appl Environ Microbiol 72:7503–7509. 10.1128/AEM.01604-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farhan Ul-Haque M, Gu W, Baral BS, DiSpirito AA, Semrau JD. 2017. Carbon source regulation of gene expression in Methylosinus trichosporium OB3b. Appl Microbiol Biotechnol 101:3871–3879. 10.1007/s00253-017-8121-z. [DOI] [PubMed] [Google Scholar]
- 19.Choi D-W, Kunz RC, Boyd ES, Semrau JD, Antholine WE, Han J-I, Zahn JA, Boyd JM, Arlene M, DiSpirito AA. 2003. The membrane-associated methane monooxygenase (pMMO) and pMMO-NADH: quinone oxidoreductase complex from Methylococcus capsulatus Bath. J Bacteriol 185:5755–5764. 10.1128/JB.185.19.5755-5764.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burrows KJ, Cornish A, Scott D, Higgins IJ. 1984. Substrate specificities of the soluble and particulate methane mono-oxygenases of Methylosinus trichosporium OB3b. Microbiology 130:3327–3333. 10.1099/00221287-130-12-3327. [DOI] [Google Scholar]
- 21.Stanley S, Prior S, Leak D, Dalton H. 1983. Copper stress underlies the fundamental change in intracellular location of methane mono-oxygenase in methane-oxidizing organisms: studies in batch and continuous cultures. Biotechnol Lett 5:487–492. 10.1007/BF00132233. [DOI] [Google Scholar]
- 22.Balasubramanian R, Smith SM, Rawat S, Yatsunyk LA, Stemmler TL, Rosenzweig AC. 2010. Oxidation of methane by a biological dicopper centre. Nature 465:115–119. 10.1038/nature08992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lieberman RL, Rosenzweig AC. 2005. Crystal structure of a membrane-bound metalloenzyme that catalyses the biological oxidation of methane. Nature 434:177–182. 10.1038/nature03311. [DOI] [PubMed] [Google Scholar]
- 24.Martinho M, Choi DW, DiSpirito AA, Antholine WE, Semrau JD, Münck E. 2007. Mössbauer studies of the membrane-associated methane monooxygenase from Methylococcus capsulatus Bath: evidence for a diiron center. J Am Chem Soc 129:15783–15785. 10.1021/ja077682b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nguyen H, Shiemke AK, Jacobs SJ, Hales BJ, Lidstrom ME, Chan SI. 1994. The nature of the copper ions in the membranes containing the particulate methane monooxygenase from Methylococcus capsulatus (Bath). J Biol Chem 269:14995–15005. 10.1016/S0021-9258(17)36565-1. [DOI] [PubMed] [Google Scholar]
- 26.Nguyen H-HT, Elliott SJ, Yip JH-K, Chan SI. 1998. The particulate methane monooxygenase from Methylococcus capsulatus (Bath) is a novel copper-containing three-subunit enzyme: isolation and characterization. J Biol Chem 273:7957–7966. 10.1074/jbc.273.14.7957. [DOI] [PubMed] [Google Scholar]
- 27.El Ghazouani A, Baslé A, Gray J, Graham DW, Firbank SJ, Dennison C. 2012. Variations in methanobactin structure influences copper utilization by methane-oxidizing bacteria. Proc Natl Acad Sci USA 109:8400–8404. 10.1073/pnas.1112921109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choi DW, Zea CJ, Do YS, Semrau JD, Antholine WE, Hargrove MS, Pohl NL, Boyd ES, Geesey GG, Hartsel SC, Shafe PH, McEllistrem MT, Kisting CJ, Campbell D, Rao V, de la Mora AM, Dispirito AA. 2006. Spectral, kinetic, and thermodynamic properties of Cu (I) and Cu (II) binding by methanobactin from Methylosinus trichosporium OB3b. Biochemistry 45:1442–1453. 10.1021/bi051815t. [DOI] [PubMed] [Google Scholar]
- 29.Chi Fru E, Gray N, McCann C, Baptista JC, Christgen B, Talbot H, Ghazouani AE, Dennison C, Graham D. 2011. Effects of copper mineralogy and methanobactin on cell growth and sMMO activity in Methylosinus trichosporium OB3b. Biogeosciences 8:2887–2894. 10.5194/bg-8-2887-2011. [DOI] [Google Scholar]
- 30.Pesch ML, Hoffmann M, Christl I, Kraemer SM, Kretzschmar R. 2013. Competitive ligand exchange between Cu–humic acid complexes and methanobactin. Geobiology 11:44–54. 10.1111/gbi.12010. [DOI] [PubMed] [Google Scholar]
- 31.Kulczycki E, Fowle DA, Kenward PA, Leslie K, Graham DW, R JA. 2007. Methanobactin‐promoted dissolution of Cu‐substituted borosilicate glass. Geobiology 5:251–263. 10.1111/j.1472-4669.2007.00102.x. [DOI] [Google Scholar]
- 32.DiSpirito AA, Semrau JD, Murrell JC, Gallagher WH, Dennison C, Vuilleumier S. 2016. Methanobactin and the link between copper and bacterial methane oxidation. Microbiol Mol Biol Rev 80:387–409. 10.1128/MMBR.00058-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim HJ, Graham DW, DiSpirito AA, Alterman MA, Galeva N, Larive CK, Asunskis D, Sherwood PM. 2004. Methanobactin, a copper-acquisition compound from methane-oxidizing bacteria. Science 305:1612–1615. 10.1126/science.1098322. [DOI] [PubMed] [Google Scholar]
- 34.Semrau JD, Jagadevan S, DiSpirito AA, Khalifa A, Scanlan J, Bergman BH, Freemeier BC, Baral BS, Bandow NL, Vorobev A, Haft DH, Vuilleumier S, Murrell JC. 2013. Methanobactin and MmoD work in concert to act as the ‘copper‐switch’ in methanotrophs. Environ Microbiol 15:3077–3086. 10.1111/1462-2920.12150. [DOI] [PubMed] [Google Scholar]
- 35.Choi DW, Do YS, Zea CJ, McEllistrem MT, Lee S-W, Semrau JD, Pohl NL, Kisting CJ, Scardino LL, Hartsel SC, Boyd ES, Geesey GG, Riedel TP, Shafe PH, Kranski KA, Tritsch JR, Antholine WE, DiSpirito AA. 2006. Spectral and thermodynamic properties of Ag (I), Au (III), Cd (II), Co (II), Fe (III), Hg (II), Mn (II), Ni (II), Pb (II), U (IV), and Zn (II) binding by methanobactin from Methylosinus trichosporium OB3b. J Inorg Biochem 100:2150–2161. 10.1016/j.jinorgbio.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 36.El Ghazouani A, Basle A, Firbank SJ, Knapp CW, Gray J, Graham DW, Dennison C. 2011. Copper-binding properties and structures of methanobactins from Methylosinus trichosporium OB3b. Inorg Chem 50:1378–1391. 10.1021/ic101965j. [DOI] [PubMed] [Google Scholar]
- 37.Semrau JD, DiSpirito AA, Obulisamy PK, Kang-Yun CS. 2020. Methanobactin from methanotrophs: genetics, structure, function and potential applications. FEMS Microbiol Lett 367:fnaa045. 10.1093/femsle/fnaa045. [DOI] [PubMed] [Google Scholar]
- 38.Kenney GE, Rosenzweig AC. 2018. Chalkophores. Annu Rev Biochem 87:645–676. 10.1146/annurev-biochem-062917-012300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gu W, Baral BS, DiSpirito AA, Semrau JD. 2017. An aminotransferase is responsible for the deamination of the N-terminal leucine and required for formation of oxazolone ring A in methanobactin of Methylosinus trichosporium OB3b. Appl Environ Microbiol 83:e02619-16. 10.1128/AEM.02619-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kenney GE, Dassama LMK, Pandelia M-E, Gizzi AS, Martinie RJ, Gao P, DeHart CJ, Schachner LF, Skinner OS, Ro SY, Zhu X, Sadek M, Thomas PM, Almo SC, Bollinger JM, Krebs C, Kelleher NL, Rosenzweig AC. 2018. The biosynthesis of methanobactin. Science 359:1411–1416. 10.1126/science.aap9437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gu W, Semrau JD. 2017. Copper and cerium-regulated gene expression in Methylosinus trichosporium OB3b. Appl Microbiol Biotechnol 101:8499–8516. 10.1007/s00253-017-8572-2. [DOI] [PubMed] [Google Scholar]
- 42.Kenney GE, Sadek M, Rosenzweig AC. 2016. Copper-responsive gene expression in the methanotroph Methylosinus trichosporium OB3b. Metallomics 8:931–940. 10.1039/c5mt00289c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nielsen AK, Gerdes K, Murrell JC. 1997. Copper‐dependent reciprocal transcriptional regulation of methane monooxygenase genes in Methylococcus capsulatus and Methylosinus trichosporium. Mol Microbiol 25:399–409. 10.1046/j.1365-2958.1997.4801846.x. [DOI] [PubMed] [Google Scholar]
- 44.Murrell JC, McDonald IR, Gilbert B. 2000. Regulation of expression of methane monooxygenases by copper ions. Trends Microbiol 8:221–225. 10.1016/s0966-842x(00)01739-x. [DOI] [PubMed] [Google Scholar]
- 45.Dassama LM, Kenney GE, Ro SY, Zielazinski EL, Rosenzweig AC. 2016. Methanobactin transport machinery. Proc Natl Acad Sci USA 113:13027–13032. 10.1073/pnas.1603578113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gu W, Haque MFU, Baral BS, Turpin EA, Bandow NL, Kremmer E, Flatley A, Zischka H, DiSpirito AA, Semrau JD. 2016. A TonB-dependent transporter is responsible for methanobactin uptake by Methylosinus trichosporium OB3b. Appl Environ Microbiol 82:1917–1923. 10.1128/AEM.03884-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Farhan Ul-Haque M, Kalidass B, Vorobev A, Baral BS, DiSpirito AA, Semrau JD. 2015. Methanobactin from Methylocystis sp. strain SB2 affects gene expression and methane monooxygenase activity in Methylosinus trichosporium OB3b. Appl Environ Microbiol 81:2466–2473. 10.1128/AEM.03981-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schalk IJ, Guillon L. 2013. Fate of ferrisiderophores after import across bacterial outer membranes: different iron release strategies are observed in the cytoplasm or periplasm depending on the siderophore pathways. Amino Acids 44:1267–1277. 10.1007/s00726-013-1468-2. [DOI] [PubMed] [Google Scholar]
- 49.Noinaj N, Guillier M, Barnard TJ, Buchanan SK. 2010. TonB-dependent transporters: regulation, structure, and function. Annu Rev Microbiol 64:43–60. 10.1146/annurev.micro.112408.134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.D'Onofrio A, Crawford JM, Stewart EJ, Witt K, Gavrish E, Epstein S, Clardy J, Lewis K. 2010. Siderophores from neighboring organisms promote the growth of uncultured bacteria. Chem Biol 17:254–264. 10.1016/j.chembiol.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miethke M, Kraushaar T, Marahiel MA. 2013. Uptake of xenosiderophores in Bacillus subtilis occurs with high affinity and enhances the folding stabilities of substrate binding proteins. FEBS Lett 587:206–213. 10.1016/j.febslet.2012.11.027. [DOI] [PubMed] [Google Scholar]
- 52.Sexton DJ, Glover RC, Loper JE, Schuster M. 2017. Pseudomonas protegens Pf‐5 favours self‐produced siderophore over free‐loading in interspecies competition for iron. Environ Microbiol 19:3514–3525. 10.1111/1462-2920.13836. [DOI] [PubMed] [Google Scholar]
- 53.Venturi V, Weisbeek P, Koster M. 1995. Gene regulation of siderophore‐mediated iron acquisition in Pseudomonas: not only the Fur repressor. Mol Microbiol 17:603–610. 10.1111/j.1365-2958.1995.mmi_17040603.x. [DOI] [PubMed] [Google Scholar]
- 54.Dean CR, Poole K. 1993. Expression of the ferric enterobactin receptor (PfeA) of Pseudomonas aeruginosa: involvement of a two‐component regulatory system. Mol Microbiol 8:1095–1103. 10.1111/j.1365-2958.1993.tb01654.x. [DOI] [PubMed] [Google Scholar]
- 55.Gensberg K, Hughes K, Smith AW. 1992. Siderophore-specific induction of iron uptake in Pseudomonas aeruginosa. J Gen Microbiol 138:2381–2387. 10.1099/00221287-138-11-2381. [DOI] [PubMed] [Google Scholar]
- 56.Koster M, van de Vossenberg J, Leong J, Weisbeek PJ. 1993. Identification and characterization of the pupB gene encoding an inducible ferric‐pseudobactin receptor of Pseudomonas putida WCS358. Mol Microbiol 8:591–601. 10.1111/j.1365-2958.1993.tb01603.x. [DOI] [PubMed] [Google Scholar]
- 57.Poole K, McKay GA. 2003. Iron acquisition and its control in Pseudomonas aeruginosa: many roads lead to Rome. Front Biosci 8:d661–d686. 10.2741/1051. [DOI] [PubMed] [Google Scholar]
- 58.Ferguson AD, Amezcua CA, Halabi NM, Chelliah Y, Rosen MK, Ranganathan R, Deisenhofer J. 2007. Signal transduction pathway of TonB-dependent transporters. Proc Natl Acad Sci USA 104:513–518. 10.1073/pnas.0609887104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moraleda‐Muñoz A, Marcos‐Torres FJ, Pérez J, Muñoz‐Dorado J. 2019. Metal‐responsive RNA polymerase extracytoplasmic function (ECF) sigma factors. Mol Microbiol 112:385–398. 10.1111/mmi.14328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schalk IJ, Yue WW, Buchanan SK. 2004. Recognition of iron‐free siderophores by TonB‐dependent iron transporters. Mol Microbiol 54:14–22. 10.1111/j.1365-2958.2004.04241.x. [DOI] [PubMed] [Google Scholar]
- 61.Kang-Yun CS, Liang X, Dershwitz P, Gu W, Schepers A, Flatley A, Lichtmannegger J, Zischka H, Zhang L, Lu X, Gu B, Ledesma JC, Pelger DJ, DiSpirito AA, Semrau JD. 2021. Evidence for methanobactin “Theft” and novel chalkophore production in methanotrophs: impact on methanotrophic-mediated methylmercury degradation. ISME J:1–10. 10.1038/s41396-021-01062-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chang J, Kim DD, Semrau JD, Lee JY, Heo H, Gu W, Yoon S. 2020. Enhancement of nitrous oxide emissions in soil microbial consortia via copper competition between proteobacterial methanotrophs and denitrifiers. Appl Environ Microbiol 87:e02301-20. 10.1128/AEM.02301-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bandow NL, Gallagher WH, Behling L, Choi DW, Semrau JD, Hartsel SC, Gilles VS, DiSpirito AA. 2011. Isolation of methanobactin from the spent media of methane-oxidizing bacteria. Methods Enzymol 495:259–269. 10.1016/B978-0-12-386905-0.00017-6. [DOI] [PubMed] [Google Scholar]
- 64.Krentz BD, Mulheron HJ, Semrau JD, Dispirito AA, Bandow NL, Haft DH, Vuilleumier S, Murrell JC, McEllistrem MT, Hartsel SC, Gallagher WH. 2010. A comparison of methanobactins from Methylosinus trichosporium OB3b and Methylocystis strain SB2 predicts methanobactins are synthesized from diverse peptide precursors modified to create a common core for binding and reducing copper ions. Biochemistry 49:10117–10130. 10.1021/bi1014375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vorobev A, Jagadevan S, Baral BS, DiSpirito AA, Freemeier BC, Bergman BH, Bandow NL, Semrau JD. 2013. Detoxification of mercury by methanobactin from Methylosinus trichosporium OB3b. Appl Environ Microbiol 79:5918–5926. 10.1128/AEM.01673-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Welander PV, Summons RE. 2012. Discovery, taxonomic distribution, and phenotypic characterization of a gene required for 3-methylhopanoid production. Proc Natl Acad Sci USA 109:12905–12910. 10.1073/pnas.1208255109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73. 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 68.Simon R. 1984. High frequency mobilization of gram-negative bacterial replicons by the in vitro constructed Tn 5-Mob transposon. Mol Gen Genet 196:413–420. 10.1007/BF00436188. [DOI] [PubMed] [Google Scholar]
- 69.Martin H, Murrell J. 1995. Methane monooxygenase mutants of Methylosinus trichosporium constructed by marker-exchange mutagenesis. FEMS Microbiol Lett 127:243–248. 10.1111/j.1574-6968.1995.tb07480.x. [DOI] [Google Scholar]
- 70.Peng P, Zheng Y, Koehorst JJ, Schaap PJ, Stams AJ, Smidt H, Atashgahi S. 2017. Concurrent haloalkanoate degradation and chlorate reduction by Pseudomonas chloritidismutans AW-1T. Appl Environ Microbiol 83:e00325-17. 10.1128/AEM.00325-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3:1101–1108. 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 72.Kalidass B, Ul-Haque MF, Baral BS, DiSpirito AA, Semrau JD. 2015. Competition between metals for binding to methanobactin enables expression of soluble methane monooxygenase in the presence of copper. Appl Environ Microbiol 81:1024–1031. 10.1128/AEM.03151-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Solovyev V, Salamov A. 2011. Automatic annotation of microbial genomes and metagenomic sequences, p 61–78. In Li RW (ed), Metagenomics and its applications in agriculture, biomedicine, and environmental studies. Nova Science Publishers, Hauppage, NY. [Google Scholar]
- 74.Im J, Lee SW, Yoon S, DiSpirito AA, Semrau JD. 2011. Characterization of a novel facultative Methylocystis species capable of growth on methane, acetate and ethanol. Environ Microbiol Rep 3:174–181. 10.1111/j.1758-2229.2010.00204.x. [DOI] [PubMed] [Google Scholar]
- 75.Knapp CW, Fowle DA, Kulczycki E, Roberts JA, Graham DW. 2007. Methane monooxygenase gene expression mediated by methanobactin in the presence of mineral copper sources. Proc Natl Acad Sci USA 104:12040–12045. 10.1073/pnas.0702879104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Muyzer G, De Waal EC, Uitterlinden AG. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700. 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1, Fig. S1 to S5. Download AEM.01793-21-s0001.pdf, PDF file, 0.9 MB (932.4KB, pdf)
Data Availability Statement
Materials and data generated in this study will be made available upon reasonable request to the corresponding author.