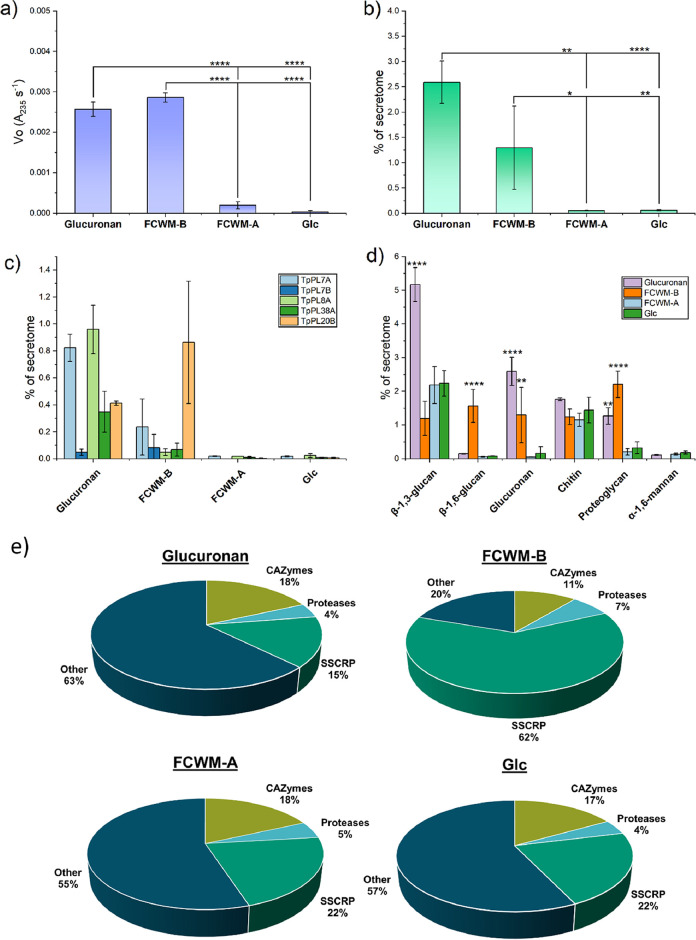
FIG 3.
Secreted proteome analysis from T. parareesei after 24 h of growth on four different carbon sources. FCWM-B = Fungal Cell Wall Medium-Basidiomycete (A. bisporus), FCWM-A = fungal cell wall medium-ascomycete (B. cinerea), Glc = Glucose. The intensity based abundances of each detected protein were normalized to the sum of each replicate. Error bars represent standard deviation of triplicate experiments. Asterisks represents P values from one-way ANOVA post hoc Tukey comparison of means. * = P ≤ 0.05, ** = P ≤ 0.01, **** = P ≤ 0.0001. a) Glucuronan assays of the fermentation supernatants under standard assay conditions. b) Summed abundances of the detected glucuronan lyases, described as percentage of the secretome for each substrate. c) Abundances of each individual glucuronan lyases. d) Abundances of CAZymes categorized by substrate specificity (Table S2). The six most abundant categories and with relevance for fungal cell wall degradation are shown. P values were calculated against the glucose fermentation only. e) Pie charts representing the averaged overall distribution of abundances in the secreted proteomes. SSCRP = Small Secreted Cysteine-Rich Proteins.