Figure 2.
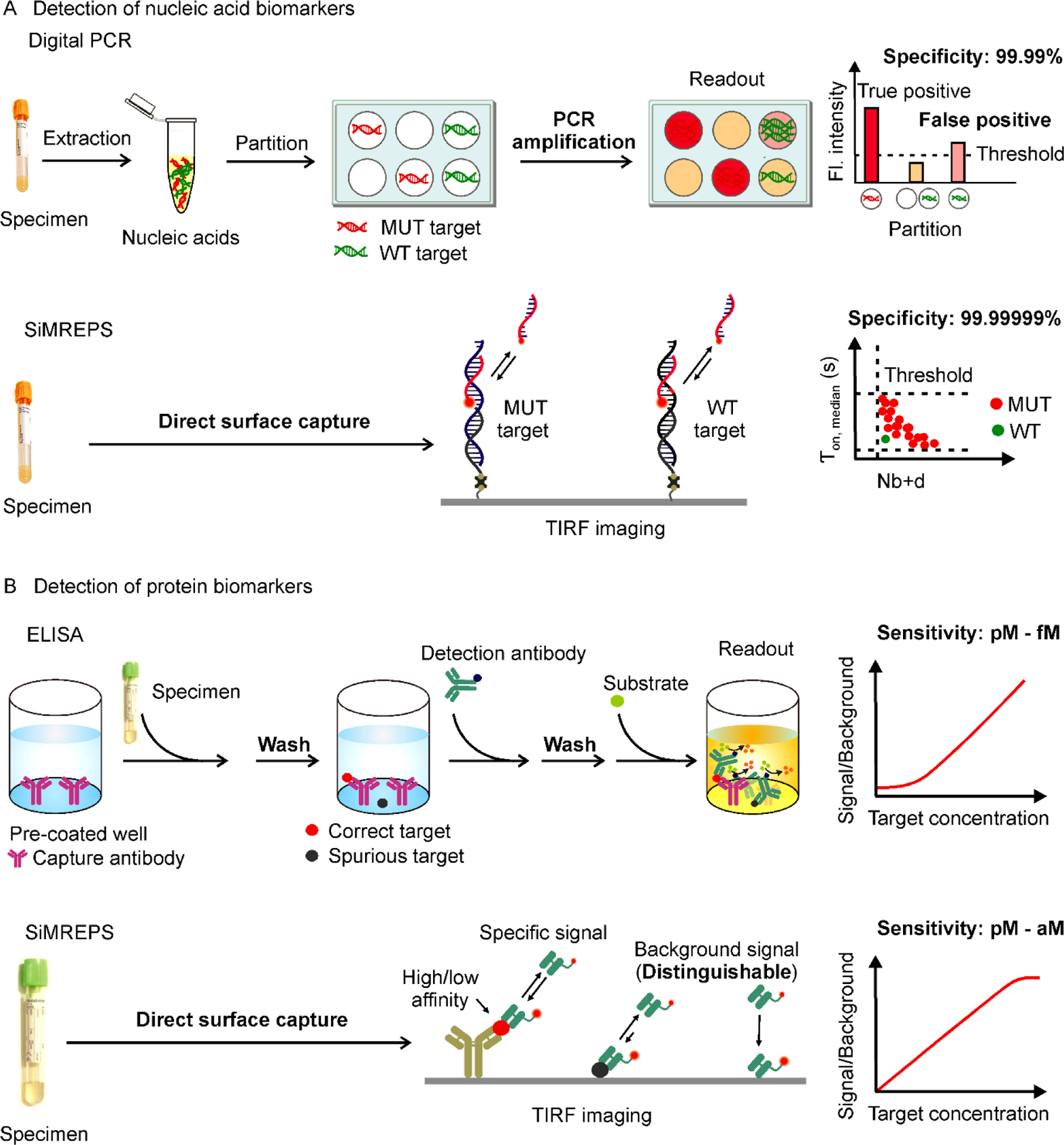
Comparison of conventional and SiMREPS approaches for the detection of nucleic acids and proteins. (A) Comparison between digital PCR and SiMREPS for detection of mutant (MUT) DNA alleles. Digital PCR is limited by its specificity due to heat-induced chemical modification of nucleobases and amplification bias that can generate false positive signals in wildtype (WT) DNA. SiMREPS is an amplification free single-molecule kinetic fingerprinting approach that utilizes transient interaction of detection probe to achieve arbitrarily high discrimination between closely related nucleic acid sequences. (B) Comparison between ELISA and SiMREPS for detection of proteins. ELISA utilizes laborious multistep stringent washing protocols, and suffers from its lower sensitivity, and dynamic range because of high background signals generated by nonspecific interaction of proteins with assay surface. SiMREPS uses a direct wash-free protocol for highly sensitive and specific detection of proteins with broader dynamic range because of its ability to suppress background signals applying kinetic thresholds.
