Figure 1. Fossil calibrated maximum likelihood phylogenetic tree of the genus Drosophila inferred from a supermatrix of 2,791 BUSCO loci (total of 8,187,056 sites).
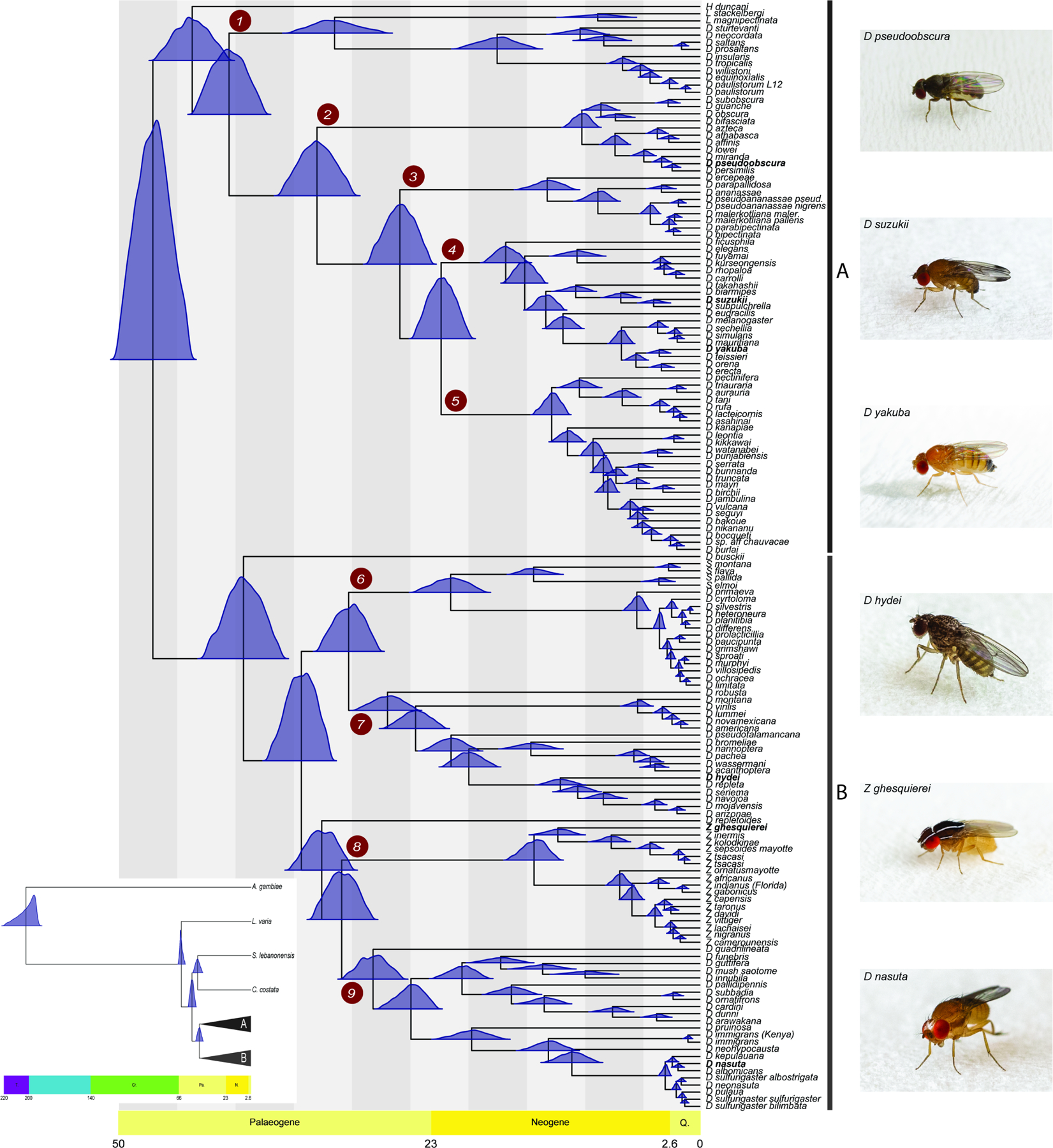
The blue distributions at each divergence point on the tree represent nodal age posterior probabilities from MCMCTree. Grauvogelia and Oligophryne fossils were used to set priors on the age of the root of the tree, Phytomyzites and Electrophortica succini were used for priors for the root of the Drosophilidae family, and Electrophortica succini and Scaptomyza dominicana were used to set priors for the crown group “Scaptomyza”, i.e. Most Recent Common Ancestor (MRCA) node of the Scaptomyza species (scheme A; Data S1). The numbered red circles denote clades for which analyses of introgression were performed. Inset: the phylogenetic and temporal relationships between our distant outgroup Anopheles gambiae, more closely related outgroup species of Drosophilidae (Leucophenga varia, Scaptodrosophila lebanonensis and Chymomyza costata), and the Drosophila genus. A and B denote the two inferred major groups within Drosophila. See also Figures S2, S5 and Data S3.
