Figure 3.
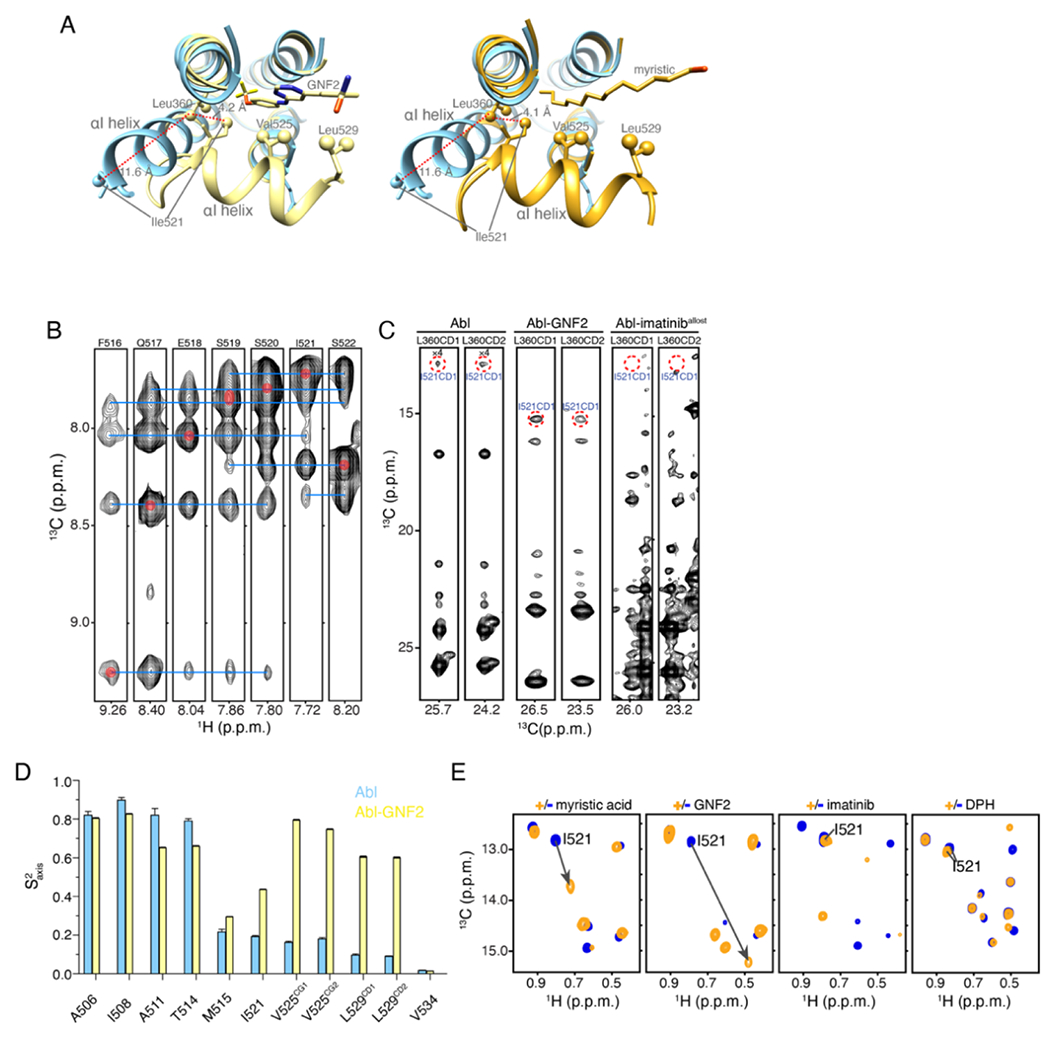
αI helix bending in Abl. (A) The structure of Abl in complex with PD173955 (blue, PDB 1M52), wherein the αI helix is not kinked, is superimposed onto the structure of Abl in complex with the allosteric inhibitor GNF2 (yellow, PDB 3K5V), which induces a kink in the αI helix (left panel) and superimposed on the structure of Abl in complex with the myristic moiety (orange, PDB 2FO0), which induces a kink in the αI-helix (right panel). (B) Strips from 3D 15N-edited 1H-1H NOESY spectra showing HN-HN NOEs that are characteristic of a helix formation. (C) Strips from 3D 13C-edited 1H-1H NOESY spectra showing the characteristic NOE between the methyl group of Leu360 and Ile521. These two residues are nearby in space in the bent conformation of the I-helix but remote in the extended αI helix conformation (panel A). (D) Methyl order parameters (S2axis) of the αI helix methyl-bearing residues in the unliganded Abl and in complex with GNF2. (E) 1H-13C methyl HMQC spectra of Abl in complex with the myristic peptide, GNF2, imatinib, and DPH. The resonance of Ile521, whose chemical shift is characteristic of the conformation of the αI helix is shown. The myristic peptide and GNF2 induce the kink to the αI helix, whereas imatinib and DPH prevent the αI helix from bending.
