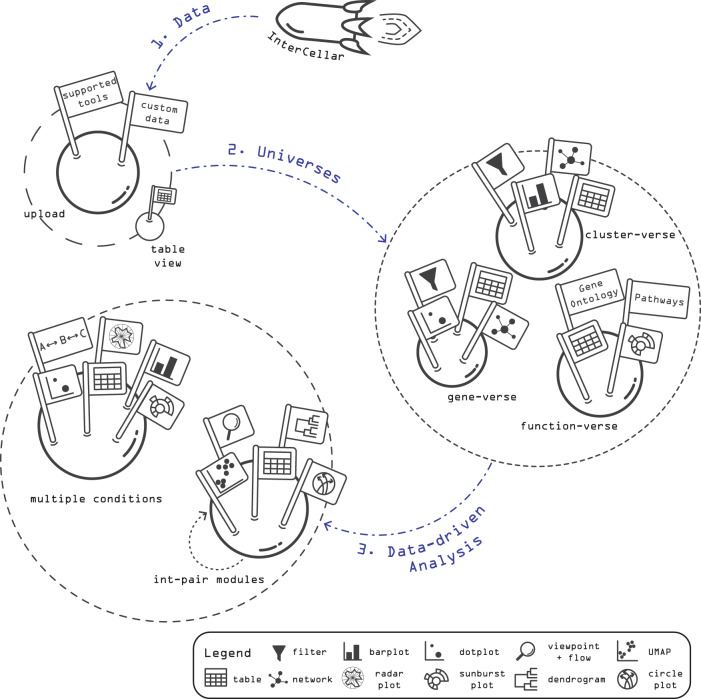
Fig. 1. Representation of InterCellar’s workflow.
The analysis workflow is composed of three main steps: (1) upload of user-provided data, containing predicted, pre-computed cell−cell interactions (CCI data); (2) exploration of three biological domains (so-called, universes), whereby the analysis focuses on cell clusters, genes, and biological functions; and (3) data-driven analysis through the definition and inspection of interaction-pair (int-pair) modules and the comparison of cell−cell communication from multiple conditions. For each step, filtering options (e.g., on cell clusters, genes, and functional databases) and multiple visualizations (e.g., networks, barplots, dotplots, circle plots, and sunburst plots) are provided to conduct a customized analysis. Moreover, all tables and figures can be saved in multiple formats, helping the interpretation of complex cell−cell interaction datasets and the finalization of publication-ready results.