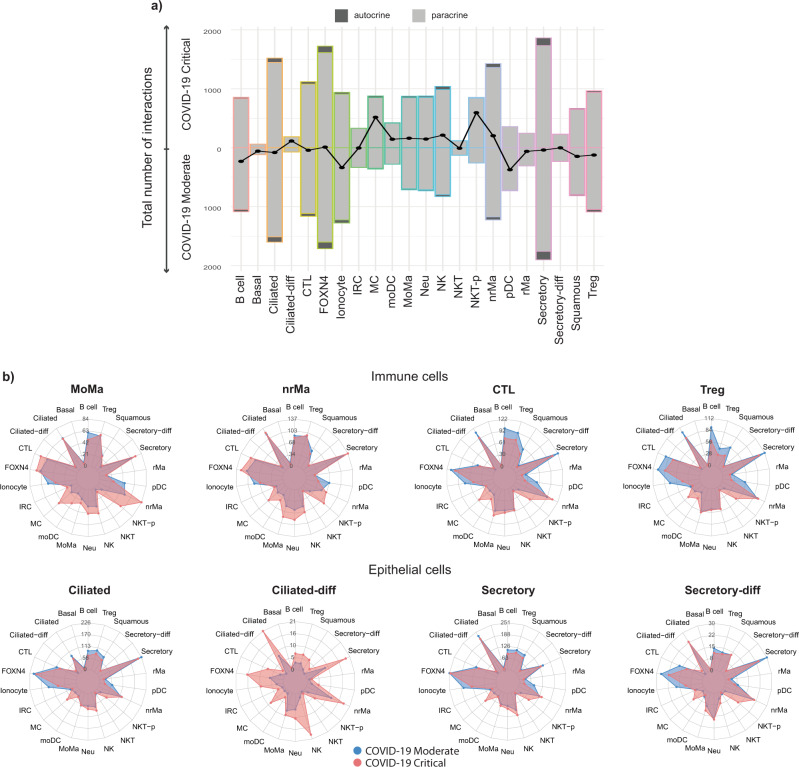
Fig. 4. InterCellar generates comprehensive insights on the number of interactions per cell cluster.
a Differences in the total number of interactions per cell type are shown in back-to-back bar plots generated by InterCellar’s multiple conditions section. All interaction flows are considered (directed-outgoing: L-R pairs, directed-incoming: R-L pairs, and undirected: L-L and R-R pairs). Dots on the black curve represent differences between the number of interactions in critical compared to moderate cases. Both autocrine and paracrine interactions are considered and plotted separately in each bar. Cell types are automatically color-coded by InterCellar for consistency throughout the analysis. See Supplementary Data 1 for source data. b Relative differences in the number of interactions are displayed in the InterCellar multiple conditions section using radar plots, comparing moderate to critical cases. All interaction flows are considered. Each plot displays the number of interactions between a specific cell type and all other cell types participating in the communication, ordered alphabetically, counter-clockwise. Cell clusters belonging to the immune cell group are represented in the first row, while epithelial cell types are shown in the second row. Cell types can be grouped in (1) epithelial cells, namely basal, ciliated, ciliated-differentiating (ciliated-diff), FOXN4+, ionocyte, IFNG responsive cell (IRC), secretory, secretory-differentiating (secretory-diff) and squamous; and (2) immune cells, namely B cell, cytotoxic T cell (CTL), regulatory T cell (Treg), natural killer (NK), natural killer T cell (NKT), natural killer T cell-proliferating (NKT-p), neutrophil (Neu), plasmacytoid dendritic cell (pDC), monocyte-derived dendritic cell (moDC), mast cell (MC), resident macrophage (rMa), non-resident macrophage (nrMa) and monocyte-derived macrophage (MoMa).