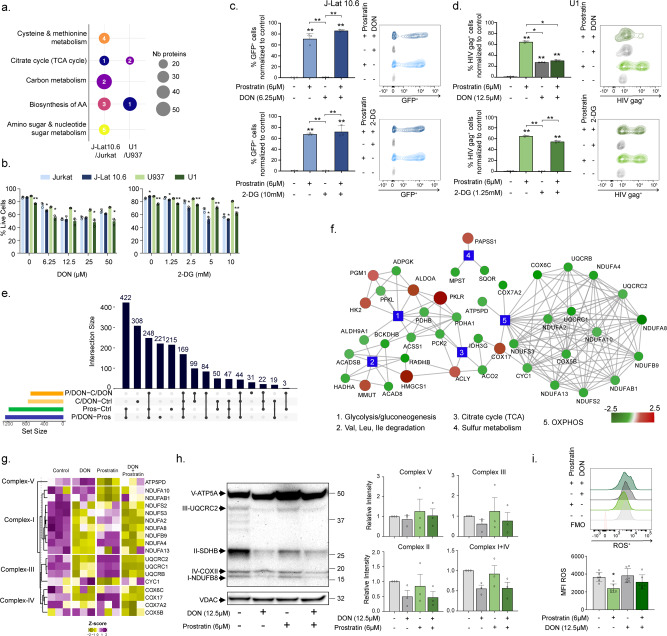
Fig. 4. Effect on cellular metabolism in latency cell models during latency reversal.
a Steady-state metabolic alterations in the HIV latency cell models, J-Lat 10.6 (dark blue) and U1 (dark green), compared to Jurkat (light blue) and U937 (light green), respectively. MSEA using the KEGG Metabolism with FDR < 0.05 is shown as bubble plots. The size of the bubble represents the number of proteins and the number, the rank of the pathway based on FDR. b Viability of latency cell models J-Lat 10.6 and U1 compared to respective parental cell lines Jurkat and U937 during 48 h DON or 2-DG treatment measured using flow cytometry. c, d HIV latency activation using prostratin (6 μM) together with DON (6.25 μM) or 2-DG (10 mM) in J-Lat 10.6 cell line (c) and DON (12.5 μM) or 2-DG (1.25 mM) in U1 cell line (d). Data represented as bar graphs (mean ± SD) of three independent experiments. Flow cytometry contour plots are from a representative sample. e The upset plot of proteins with differential abundance between control vs. DON (C/DON-Ctrl, yellow), control vs. prostratin (Pros-Ctrl, green), prostratin vs. DON + prostratin (P/DON-Pros, blue), and DON + prostratin vs. DON (P/DON-C/DON, orange) in U1 cells corrected for U937 cells. Horizontal bars show the number of proteins found in each comparison. Vertical bars display intersects between comparisons as indicated in the matrix below the graph. f Network of the proteins differing significantly between control U1 and DON treated U1 (LIMMA, FDR < 0.1, n = 758). Blue colored rectangular nodes represent KEGG pathways and colored circles represent proteins. The color gradient was applied depending on log2FC for each metabolite from green (decreased in U1-DON) to red (increased in U1-DON). The size of the bubble is also proportional to log2FC. g Heatmap showing OXPHOS proteins levels in U1 treated with DON and prostratin. Proteins were selected based on comparison U1 vs U1-DON (LIMMA, FDR < 0.1) and their association with the OXPHOS KEGG pathway. Proteins were separated based on their complexes (from I to V). h Western blot analysis and quantification of OXPHOS proteins during DON treatment in U1 cells with a representative blot of three independent experiments (Fig. S10) are shown here. Quantification of western blot represented as bar graphs with mean ± SEM. i Measurement of ROS in latency cell model U1 during prostratin and DON treatments. Data represented as mean ± SD of three independent experiments All statistical analysis was performed using unpaired t-test or Mann–Whitney U-test (*p < 0.05, and **p < 0.001).