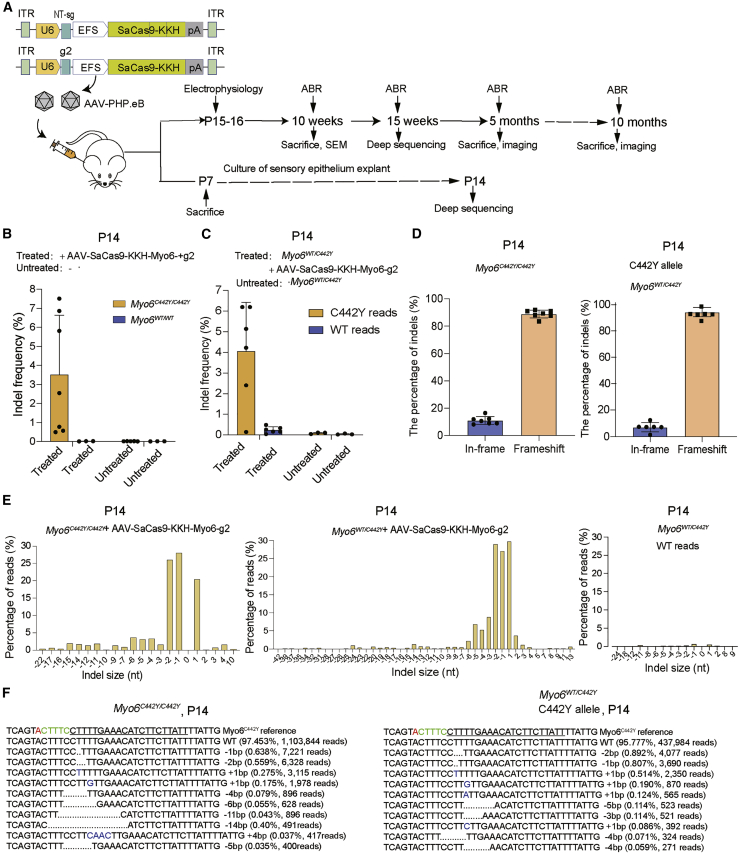
Figure 2.
In vivo genome editing with the AAV.PHP.eB-SaCas9-KKH-Myo6-g2 system
(A) Schematic of the AAV vectors (control and g2) and the experimental overview of the in vivo studies. (B and C) The indel percentages at P14 in Myo6C442Y/C442Y, Myo6WT/C442Y, and Myo6WT/WT in the sensory epithelia were determined by targeted NGS. The samples were from ears treated with or without AAV.PHP.eB-SaCas9-KKH-Myo6-g2 at P0–2 via the scala media (n = 3–7). Data are presented as the mean ± SD. (D) The indels causing in-frame versus frameshift mutations were calculated after AAV.PHP.eB-SaCas9-KKH-Myo6-g2 infection in Myo6C442Y/C442Y and Myo6WT/C442Y mice. (E) Indel profiles from SaCas9-KKH-Myo6-g2-transfected Myo6C442Y/C442Y and Myo6WT/C442Y mice. Negative numbers represent deletions, and positive numbers represent insertions. Myo6C442Y and Myo6WT reads are plotted separately. Sequences without indels (value = 0) are omitted from the chart. (F) The most abundant reads in the AAV.PHP.eB-SaCas9-KKH-Myo6-g2-treated Myo6C442Y/C442Y and Myo6WT/C442Y mice are shown.