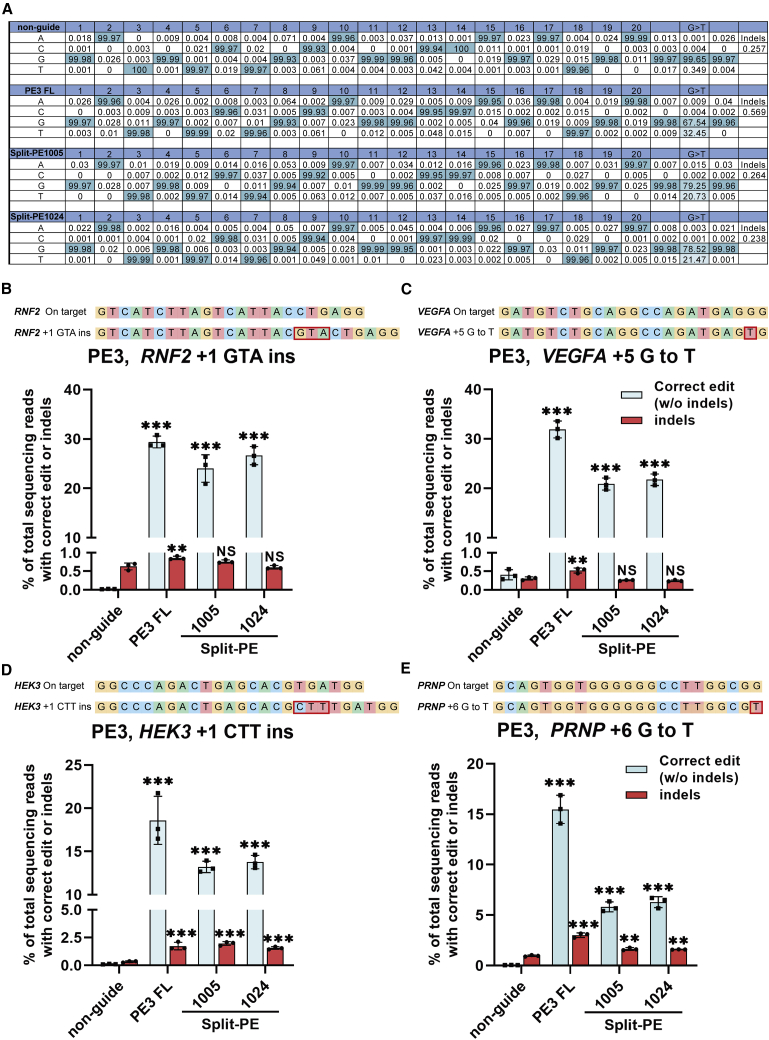
Figure 2.
Prime editing of endogenous sites in HEK293T cells by split-PEs
(A) Example of deep-sequencing data analysis of G-to-T conversion at endogenous sites targeted by split-PEs. Single-nucleotide transversion editing efficiency was analyzed by MATLAB, whereas insertion editing efficiency and indel frequency were analyzed by CRISPResso2. (B) Targeted insertion of GTA at RNF2 in HEK293T. (C) Targeted transversion of G to T at VEGFA in HEK293T. (D) Targeted insertion of CTT at HEK3 in HEK293T. (E) Targeted transversion of G to T at PRNP in HEK293T. Editing efficiencies represent sequencing reads that contain the correct edit and do not contain indels among total sequencing reads. Indels were also plotted for comparison. non-guide, HEK293T cells transfected with full-length PE2 but not pegRNA. PE3 FL, HEK293T cells transfected with full-length PE2, pegRNA, and nick-gRNA. Values and error bars represent the mean ± SD of three independent biological repeats. One-way ANOVA was used for calculating statistical significance (∗∗p < 0.01; ∗∗∗p < 0.001).