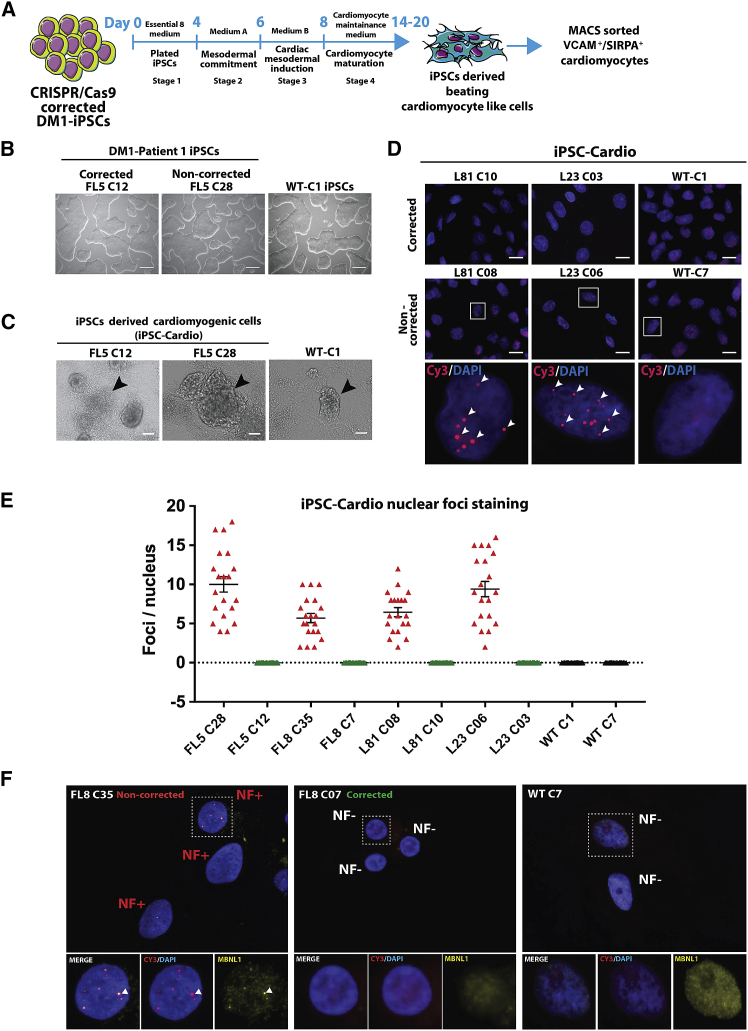
Figure 1.
Cardiomyogenic differentiation of CRISPR-Cas9 RNP-corrected and non-corrected DM1 induced pluripotent stem cells (DM1-iPSCs)
(A) Schematic representation of the various stages of the generation of iPSC-derived cardiomyogenic cells from human iPSCs. The flowchart also depicts the step of magnetic activated cell sorting (MACS) of VCAM+ and/or SIRPA+ cells after differentiation. (B) Representative phase contrast images of DM1-iPSC clonal lines (DM1 patient FL: FL5 C12 and FL5 C28) obtained after CRISPR-Cas9 treatment and clonal isolation. A healthy wild-type (WT) iPSC line (WT-C1) cultured under feeder-free conditions is also shown (scale bars, 250 μm). (C) Representative phase contrast images of DM1 and WT iPSC-derived cardiomyogenic differentiation cells (iPSC-Cardio) obtained after day 14 of differentiation. The cardiomyogenic beating cell clusters are dense multi-layered structures marked with black arrowheads (scale bars, 100 μm). (D) Representative image of DM1 cells and iPSC-Cardio stained for RNA foci by fluorescence in situ hybridization (FISH). Cardiomyogenic cells from CRISPR-Cas9-corrected DM1-iPSC clones (DM1 patient L: L81-C10 and L23-C03) and non-corrected DM1-iPSC clones (DM1 patient L: L81-C08 and L23-C06) were analyzed. Healthy WT iPSC line-derived cardiomyogenic cells (WT-C1) were taken as the negative control for FISH staining. An antisense Cy3-labeled probe was used against trinucleotide CUGexp RNA. Selected nuclei from the non-corrected samples were zoomed into (bottom panel) to show the ribonuclear foci (red) indicated by arrowheads. Upper panel represents stained nuclei at lower magnification (scale bars, 20 μm), and the bottom-most panel represents higher magnification of the selected region. Nuclei were counterstained with DAPI. (E) Graph representing foci per nucleus for cardiomyocytes derived from CRISPR-Cas9-corrected and non-corrected DM1-iPSCs. WT iPSC-derived cardiomyocytes were taken as negative controls. The foci per nucleus were calculated and are represented by each data point in the graph. The data are represented as mean ± SEM. (F) Dual staining for MBNL1 protein and analysis of localization with a DMPK mutated transcript with CTGexp repeats in the ribonuclear foci complex in the corrected FL8-C7 DM1-iPSC- and non-corrected FL8-C35 DM1-iPSC-derived cardiac cells. WT cells (WT-C7) were also stained as a healthy control. Each representative image is a maximum intensity z projection of the z slices. Enlarged views of selected ribonuclear foci negative (NF−) and positive (NF+) nuclei are represented under different filters. Nuclei were counterstained with DAPI.