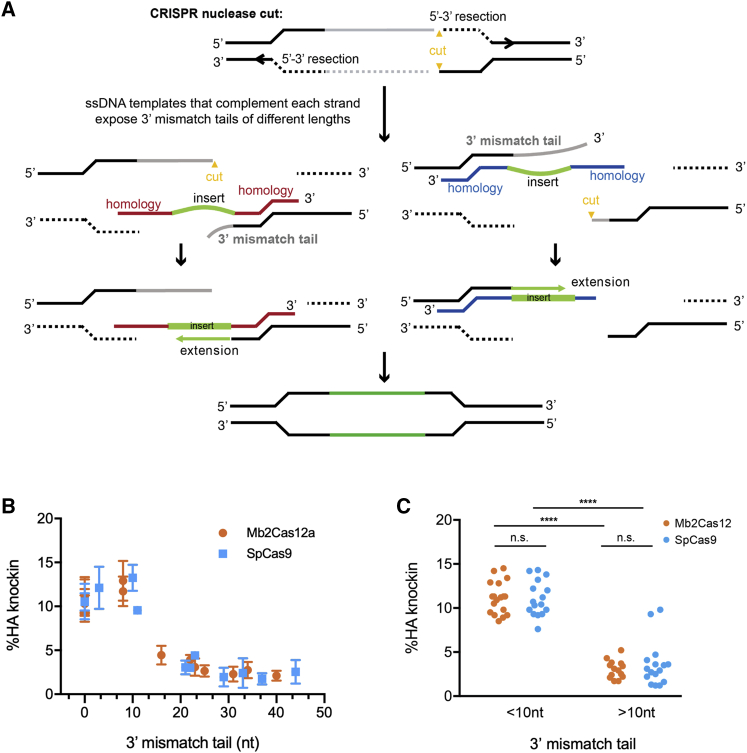
Figure 3.
The length of the 3′ mismatch tail determines replacement efficiency with short single-stranded HDRT
(A) A model showing where a 3′ mismatch tail occurs. A cut site (yellow) is introduced into a region of the gene targeted for replacement (gray), asymmetrically dividing this region. Efficient 5′ to 3′ resection exposes two 3′ ends. An HDRT can complement a strand with a short (left figures) or long 3′-mismatch tail (right figures), which must be removed before the remaining 3′ end can be extended to complement the HDRT insert region and its distal homology arm. We propose that the removal of this 3′ mismatch tail is a rate-limiting step determining editing efficiency when genomic sequences are replaced. (B) The predicted length of the 3′-mismatch tail in experiments using HDRT-A and HDRT-B presented in Figure 2 are plotted against the efficiency with which an HA tag is introduced into the HCDR3 region, as determined by flow cytometry. Error bar indicates SEM from two independent experiments. (C) A comparison of editing efficiency between those with short (<10 nt) or long (>10 nt) 3′ mismatch tails. Editing by SpCas9 or Mb2Cas12a is significantly more efficient with short 3′ mismatch tails, as determined by one-way ANOVA with Tukey's multiple comparison test (p < 0.0001). Dots represent pooled data from two independent experiments.