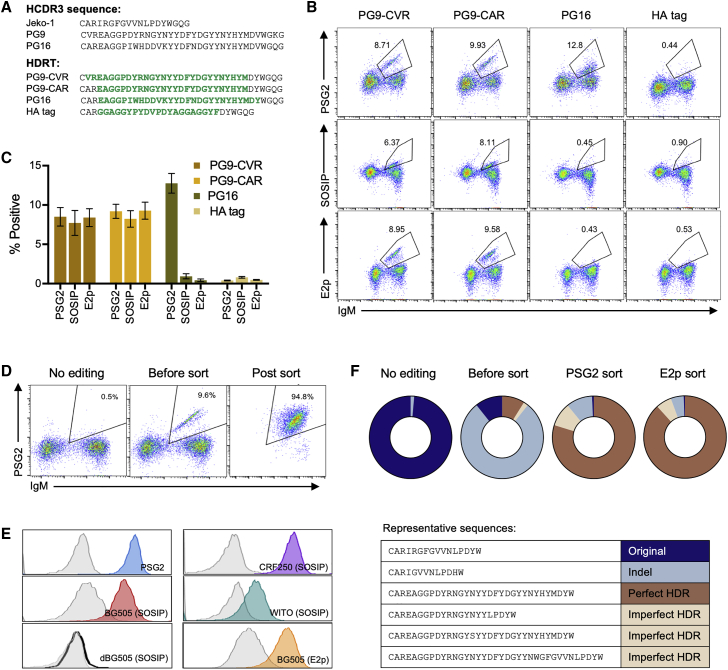
Figure 4.
The BCR specificity of Jeko-1 cells can be reprogrammed with a novel HCDR3
(A) The amino acid sequence of the native Jeko-1 cell HCDR3 region and those of the HIV-1 neutralizing antibodies PG9 and PG16 are shown. The amino acid translations of the new sequences from the four HDRTs used in the subsequent panels are represented in green italics, in the context of the remaining Jeko-1 region. (B) Mb2Cas12a RNP targeting the GTTC PAM of site 4 in Jeko-1 cells shown in Figure 2B were co-electroporated with the indicated HDRT. Editing efficiency was monitored on the vertical axis by flow cytometry with fluorescently labeled PSG2, an antibody that recognizes sulfotyrosines within the PG9 and PG16 HCDR3 region, a similarly labeled HIV SOSIP or E2p. The horizontal axis indicates IgM expression, and its loss indicates imprecise NHEJ after Mb2Cas12a-mediated cleavage. Note that introduction of a PG16 HCDR3 was efficient, as indicated by PSG2 recognition, but unlike the PG9 HCDR3, it did not bind the Env trimer. Cells edited to express an HA tag did not bind any reagent. SOSIP proteins were derived from the BG505 HIV-1 isolate. (C) A summary of three independent experiments similar to that shown in (B) of flow cytometric studies used to generate (B). Error bars indicate SD. (D) Jeko-1 edited with PG9-CAR HDRT were enriched by FACS with the anti-sulfotyrosine antibody PSG2. (E) Cells enriched in (D) were analyzed 2 weeks later by flow cytometry for their ability to bind PSG2, a BG505-based nanoparticle (BG505-E2p), SOSIP trimers derived from the indicated HIV-1 isolate, or an V2 apex negative mutant (dBG505-SOSIP). Gray control indicates wild-type Jeko-1 cells. (F) Unedited Jeko-1 cells and those edited with PG9-CAR HDRT without sorting, or sorted with PSG2 or with E2p, were analyzed by NGS of the VDJ region. Sequences were divided into four categories, depending on whether the edited sequence exactly matched the HDRT (Perfect HDR), whether the HDRT sequence was visible but modified (Imperfect HDR), whether the original Jeko-1 HCDR3 region was intact (Original), or whether this region was modified by NHEJ as indicated by the presence of insertions or deletions (Indel). Representative examples of each category are shown below the charts.