Figure 5.
Editing primary human B cells with HDRT recognizing consensus sequences of multiple VH families
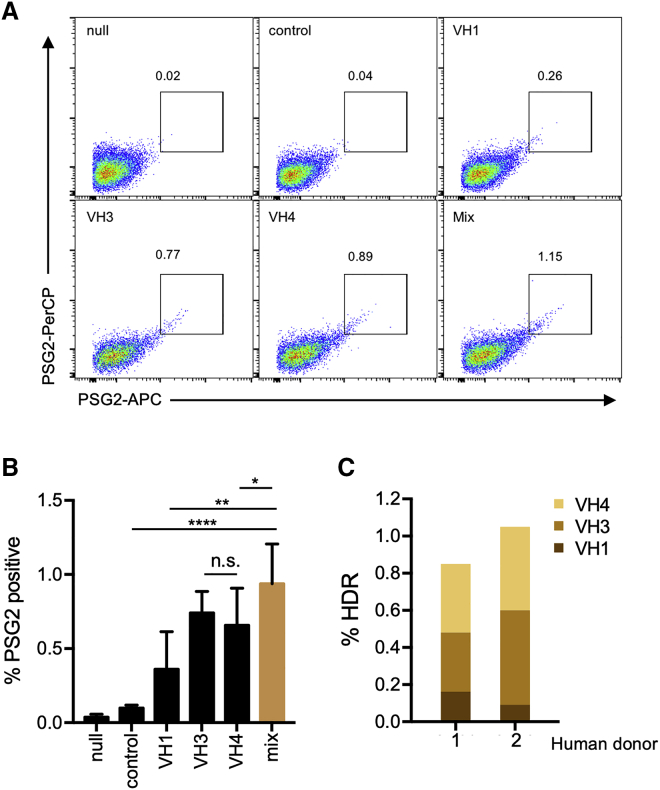
(A) A panel of PG9-CAR HDRT with homology arms complementary to JH4 and to consensus VH1-, VH3-, and VH4-family sequences were evaluated for their ability to edit primary human B cells. Cells electroporated with Mb2Cas12a RNP and PG9-CAR HDRT were analyzed by flow cytometry with the anti-sulfotyrosine antibody PSG2 modified with two distinct fluorophores to eliminate non-specific binding from either fluorophore. (B) A summary of results from experiments similar to that shown in (A), using primary B cells from three independent donors. Note that a mixture of three HDRTs edited more cells than any individual HDRT. Null indicates that cells were not electroporated and control indicates cells electroporated with Mb2Cas12a RNP and an HDRT that is not homologous to any sequence in the human genome. Mix indicates cells electroporated with RNP and an equimolar mixture of HDRT with VH1-, VH3-, and VH4-specific homology arms. Error bars indicate range of three independent experiments, and asterisks indicate statistical significance calculated by one-way ANOVA with Tukey's multiple comparison test (∗p < 0.5; ∗∗p < 0.01; ∗∗∗∗p < 0.0001). (C) NGS analysis of primary B cells from two human donors. %HDR was quantified as described in Figure 4F, including both perfect and imperfect. The portion of VH-family of edited cells in HDR-positive sequences was also counted.