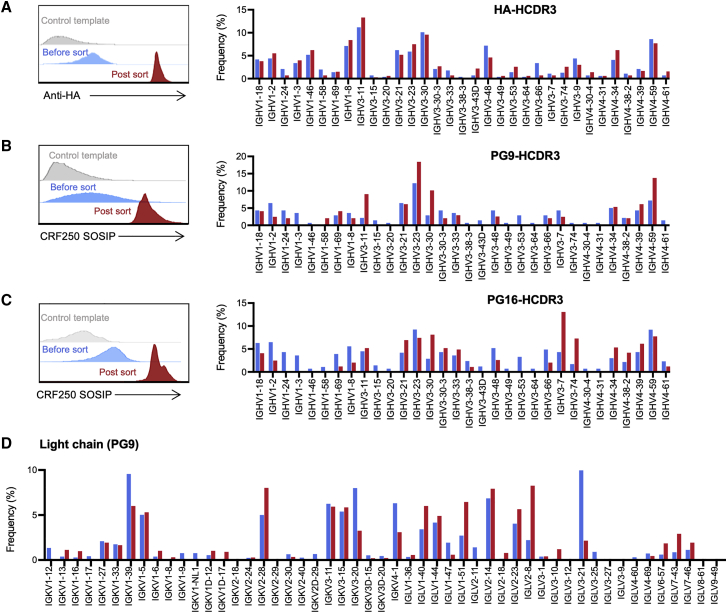
Figure 6.
Reprogrammed primary human B cells retain VH-gene and light-chain diversity
Primary cells were electroporated with Mb2Cas12a RNP and HDRT encoding an HA tag (A) or the HCDR3 regions of the HIV-1 neutralizing antibodies PG9 (B) and PG16 (C), with the same mixture of homology arms as those used in Figure 5. Cells were sorted with an anti-HA antibody (HA tag [A]) or an SOSIP trimer derived from the CRF_AG_250 isolate (B and C). Approximately 2% to 3% of cells were sorted by the SOSIP trimer. Edited cells were analyzed by NGS before and after sorting, the percentages of PG9 and PG16-HCDR3 inserts were enriched from ∼1% to 30%. The frequency of each VH1-, VH3-, and VH4-family genes from BCR sequences bearing the desired inserts was measured. Flow cytometry histograms display one of two experiments with similar results, and bar graphs indicate the mean of those two experiments. (D) The frequency of each light chain genes of PG9-HCR3 inserted primary cells pre (blue) and post (red) sorting was analyzed by NGS. Bar graphs indicate the mean of two independent experiments.