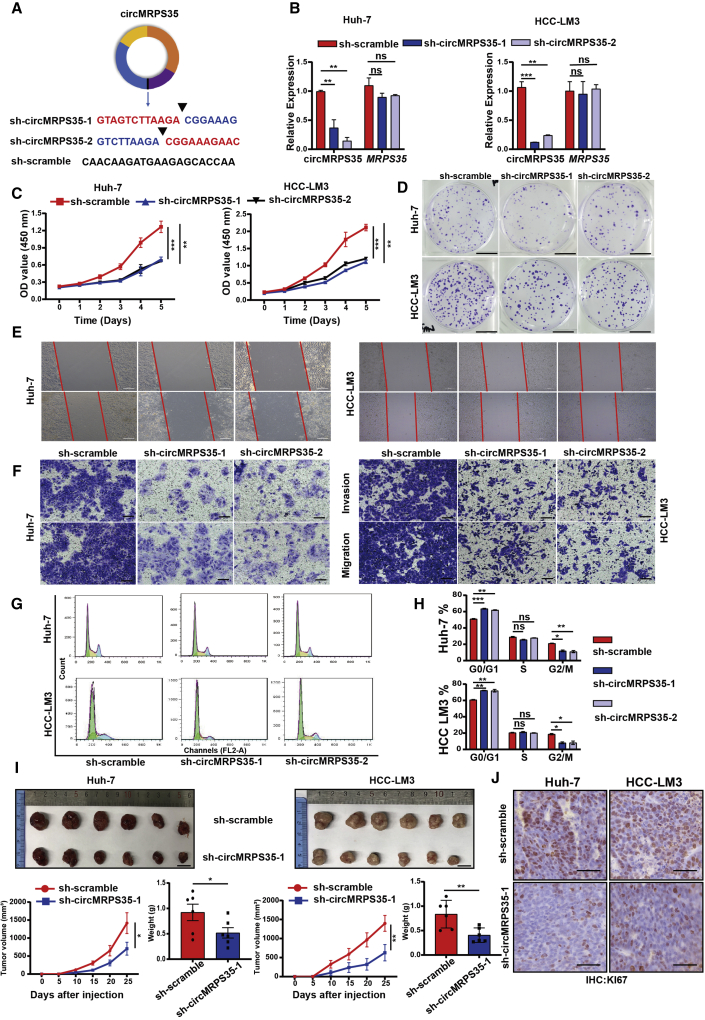
Figure 2.
circMRPS35 acts as an oncogene in HCC cells
(A) Schematic representation of target sequences of shRNAs of circMRPS35. (B) Quantitative real-time PCR analysis of circMRPS35 and MRPS35 of circMRPS35 KD Huh-7 and HCC-LM3 cells. (C) Cell viability assays were used to test proliferation of circMRPS35 KD or control Huh-7 and HCC-LM3 cells. (D) Colony formation assays were performed to test cell growth of circMRPS35 KD or control Huh-7 and HCC-LM3 cells (scale bars, 1 cm). (E) Wound-healing experiments were used to detect cell migration of circMRPS35 KD or control Huh-7 and HCC-LM3 cells (scale bars, 100 μm). (F) Transwell assays of invasion and migration in circMRPS35 KD or control Huh-7 and HCC-LM3 cells (scale bars, 100 μm). (G and H) Cell cycle assays were used to detect cell cycle arrest levels of circMRPS35 KD or control Huh-7 and HCC-LM3 cells: G0/G1, green only; S, yellow; G2/M, blue only. (I) BALB/c nude mice (n = 6 each group) were injected sh-circMRPS35-1 or sh-scramble Huh-7 and HCC-LM3 cells. Sizes of xenograft tumors were measured every 5 days, and weights of xenograft tumors were summarized after animals were sacrificed (scale bars, 1 cm). (J) IHC analysis of Ki67 for sh-circMRPS35-1 or sh-scramble Huh-7 and HCC-LM3 xenograft tumors tissues (scale bars, 100 μm). Error bars represent the means ± SEM of three independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.