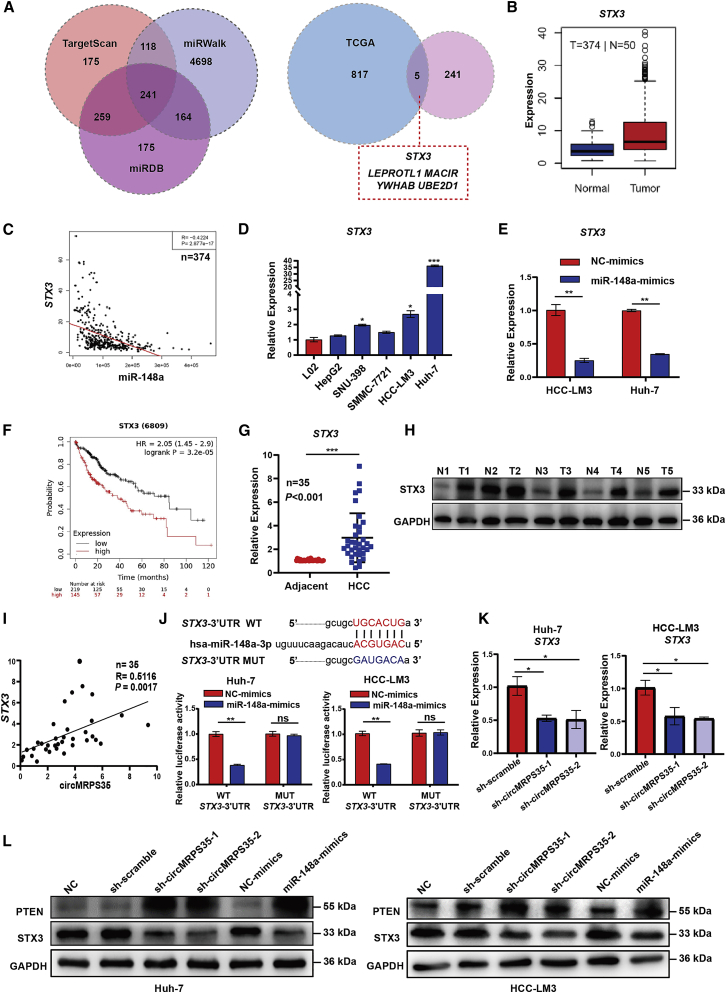
Figure 4.
circMRPS35 sponges miR-148a to regulate the expression of STX3 in HCC cells
(A) Schematic illustration showing the target mRNAs of miR-148a predicted by overlapping TargetScan, miRWalk and miRDB databases (left) and the HCC TCGA database (right). (B and C) TCGA analysis of expression of STX3 in HCC tissues and correlation analysis of miR-148a and STX3 expression. (D) Quantitative real-time PCR assays of STX3 expression in HCC cell lines compared to L02 cells. (E) Quantitative real-time PCR assays of STX3 in miR-148a overexpression HCC-LM3 and Huh-7 cells. (F) Kaplan-Meier analysis of the expression of STX3 in HCC (n = 364). (G) Quantitative real-time PCR analysis of STX3 in 35 pairs of HCC and adjacent tissues (n = 35). (H) Western blot analysis of STX3 in five pairs of HCC and adjacent tissues. (I) Correlation analysis of circMRPS35 and STX3 expression (n = 35). (J) Predicted complementary binding sites between STX3 and miR-148a (upper); a luciferase reporter assay was used to test the binding of STX3 and miR-148a in Huh-7 and HCC-LM3 cells (lower). (K) Quantitative real-time PCR assays of STX3 in circMRPS35 KD or control Huh-7 and HCC-LM3 cells. (L) Western blot analysis of STX3 and PTEN in circMRPS35 KD and miR-148a overexpression and control Huh-7 and HCC-LM3 cells. Error bars represent the means ± SEM of three independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.