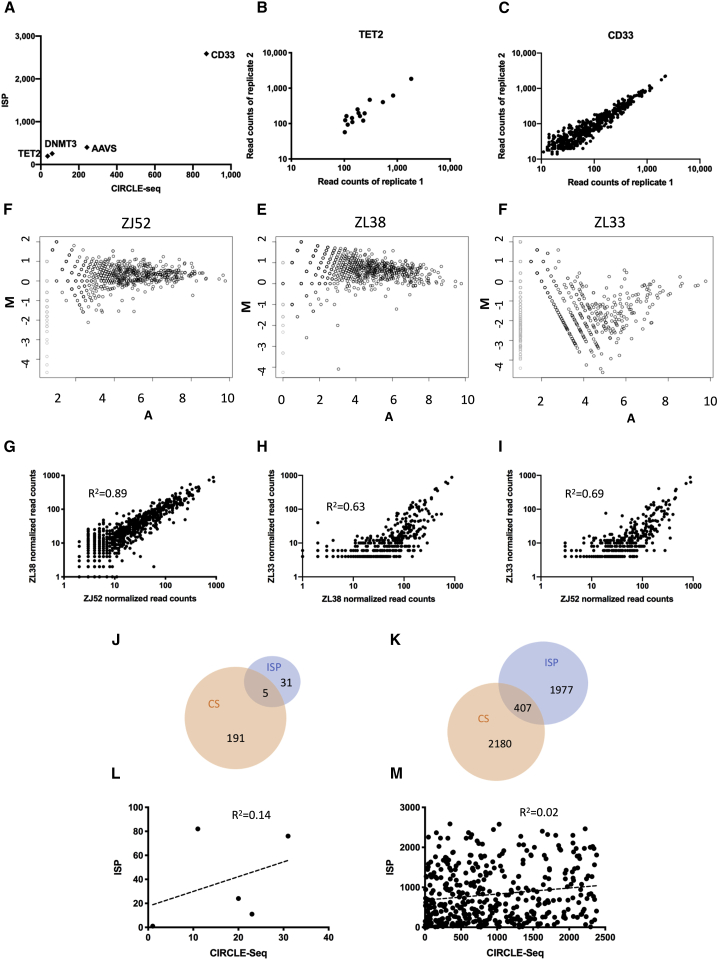
Figure 1.
CIRCLE-seq reproducibility and comparison of retrieved sites to ISP
(A) Number of sites predicted by CIRCLE-seq versus ISP for 4 different gRNAs on a log scale. (B) Normalized read counts retrieved via TET2 gRNA CIRCLE-seq technical replicates performed on the same RM blood DNA sample are shown. (C) Normalized read counts retrieved via CD33 gRNA CIRCLE-seq technical replicates performed on the same RM blood DNA sample are shown. Pearson correlation R2 values are shown. (D–F) Visualization of differences between the sites predicted by CIRCLE-seq for the CD33 gRNA performed on blood DNA samples from animals ZJ52, ZL38, and ZL33, respectively, with an MA plot, where M is the log ratio scale and A is the mean average scale is shown (average read count for each predicted site calculated using all 3 sets of data). (G–I) Normalized read count plots (log scale) show the correlation between the reads for the CD33 gRNA off-target sites detected by CIRCLE-seq, 2 animals at a time, with pairwise Pearson correlations shown for each comparison. (J) Overlap between TET2 sites predicted with CIRCLE-seq on ZL26 DNA and by ISP is shown. (K) Overlap between CD33 sites predicted by CIRCLE-seq on DNA from ZL33, ZL38, and ZJ52 and by ISP is shown. (L and M) Plot of the ranks for off-target sites predicted by CIRCLE-seq versus ISP and Spearman correlations between the two rankings for the TET2 gRNA (L) and the CD33 gRNA (M) are shown.