Figure 1.
Test and validation of an effective sgRNA to knock down the HBB gene
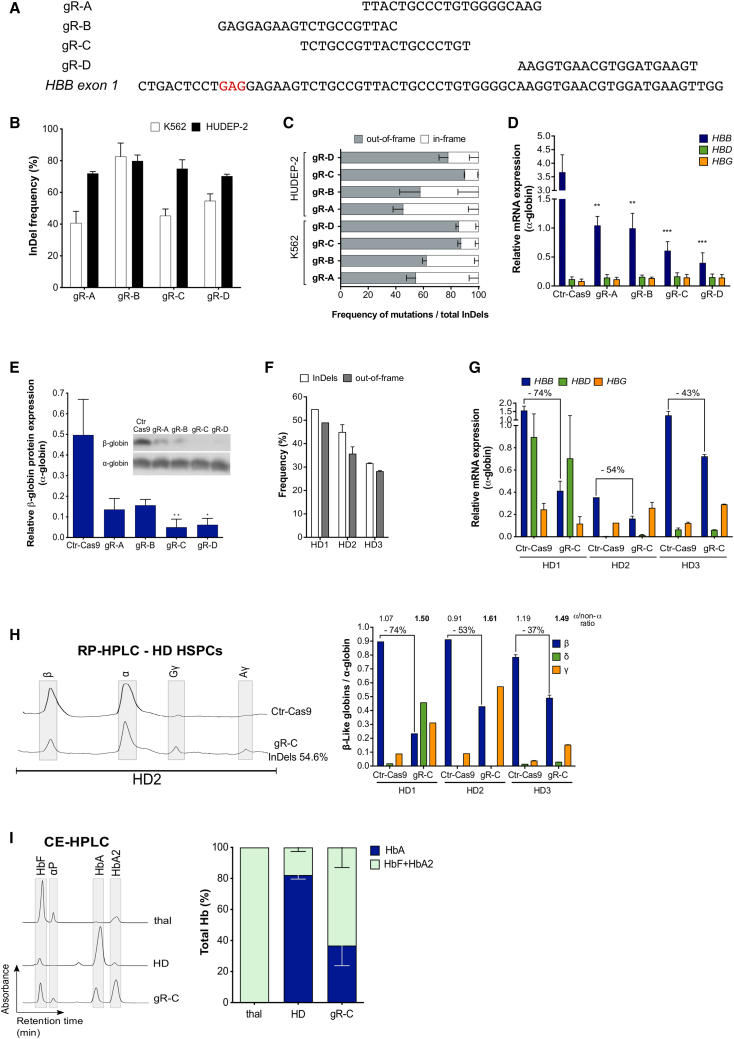
(A) sgRNAs are aligned to their complementary on-target loci on exon 1 of the HBB gene. gR-B targets a region containing the SCD mutation, and therefore its sequence was modified to target the WT HBB gene during the validation experiments in K562 and HUDEP-2 erythroid cell lines, and in HD HSPCs. The codon containing the SCD mutation is highlighted in red. (B) Editing efficiency by TIDE analysis after PCR amplification of the target region and Sanger sequencing in Cas9-GFP+ K562 (n = 3 for gR-A, 4 for gR-B, 6 for gR-C, and 5 for gR-D) and HUDEP-2 (n = 5 for gR-C; n = 4 for gR-A, gR-B, and gR-D) erythroid cell lines. Transfection efficiency was 55%–70% for K562 and 30%–60% for HUDEP-2. Data are expressed as mean ± SEM. (C) Out-of-frame mutations identified using TIDE after PCR amplification of the target region and Sanger sequencing in K562 (n = 3 for gR-A, 4 for gR-B, 6 for gR-C, and 5 for gR-D) and HUDEP-2 (n = 5 for gR-C; n = 4 for gR-A, gR-B, and gR-D) erythroid cell lines. Data are expressed as mean ± SEM. The frequency of out-of-frame mutations for gR-C was significantly higher than for gR-A (p < 0.0001 in K562 and p < 0.05 in HUDEP-2) and gR-B (p < 0.005 in K562). In K562, the frequency of out-of-frame mutations for gR-D was significantly higher than for gR-A and gR-B (p < 0.0005). In both K562 and HUDEP-2, no differences were observed between gR-C and gR-D (one-way ANOVA test: Tukey’s multiple comparisons test). (D) Relative expression of HBB, HBD, and HBG mRNAs normalized to α-globin, detected by qRT-PCR in HUDEP-2 cells. Data are expressed as mean ± SEM (n = 3). ∗∗p < 0.01 between gR-A or gR-B versus control (Ctr)-Cas9; ∗∗∗p < 0.001 between gR-C or gR-D versus Ctr-Cas9 (two-way ANOVA test: Dunnett’s multiple comparisons test). Ctr-Cas9 samples are HUDEP-2 cells transfected only with the Cas9-GFP plasmid. (E) Relative expression of the β-globin chain normalized to the α-globin chain, as measured by western blot analysis in differentiated HUDEP-2 cells. Data are expressed as mean ± SEM (n = 4 for gR-C; n = 3 for gR-A, gR-B, and gR-D). ∗p < 0.05 and ∗∗p < 0.01 versus Ctr-Cas9 (two-way ANOVA test: Sidak’s multiple comparisons test). A representative western blot image is shown. Ctr-Cas9 samples are HUDEP-2 cells transfected only with the Cas9-GFP plasmid. (F) Editing efficiency (indels) and frequency of out-of-frame mutations measured by TIDE analysis after PCR amplification of the target region and Sanger sequencing in G-CSF-mobilized peripheral blood (mPB) HD HSPCs edited with gR-C (n = 1 for HD1, n = 2 for HD2, and n = 2 for HD3). Data are expressed as mean ± SEM. (G) Relative expression of HBB, HBD, and HBG mRNAs normalized to α-globin, measured by qRT-PCR in erythroid cells derived from G-CSF-mPB HD HSPCs transfected with Cas9-GFP plasmid only (Ctr-Cas9) or with both Cas9-GFP and gR-C plasmids (gR-C). HBB decrease upon gR-C treatment is reported as percentage above the histogram bars (n = 1 for HD1, n = 2 for HD2, and n = 2 for HD3). Data are expressed as mean ± SEM. (H) RP-HPLC quantification of globin chains in erythroid cells derived from G-CSF-mPB HD HSPCs transfected with Cas9-GFP plasmid only (Ctr-Cas9) or with both Cas9-GFP and gR-C plasmids (gR-C). β-Like globin chains are normalized over α-globin chains (n = 1 for HD1, n = 2 for HD2, and n = 2 for HD3; n = 5 biological replicates). The percentage of β-globin decreases upon gR-C treatment and the α-globin/non-α-globin ratio is reported above the histogram bars. Data are expressed as mean ± SEM. A representative chromatogram is reported in the left panel. (I) CE-HPLC analysis of Hb tetramers in β-thalassemic cells (thal) and in erythroid cells derived from G-CSF-mPB HD HSPCs treated or not with gR-C. gR-C-treated samples have a hemoglobin expression pattern similar to the profile observed in in vitro-differentiated erythroid cells from a β-thalassemia patient. We calculated the percentage of Hb types over the total Hb tetramers (n = 2 donors). CE-HPLC chromatograms are reported on the left panel. αp, α-precipitates. Data are expressed as mean ± SEM.