Figure 2.
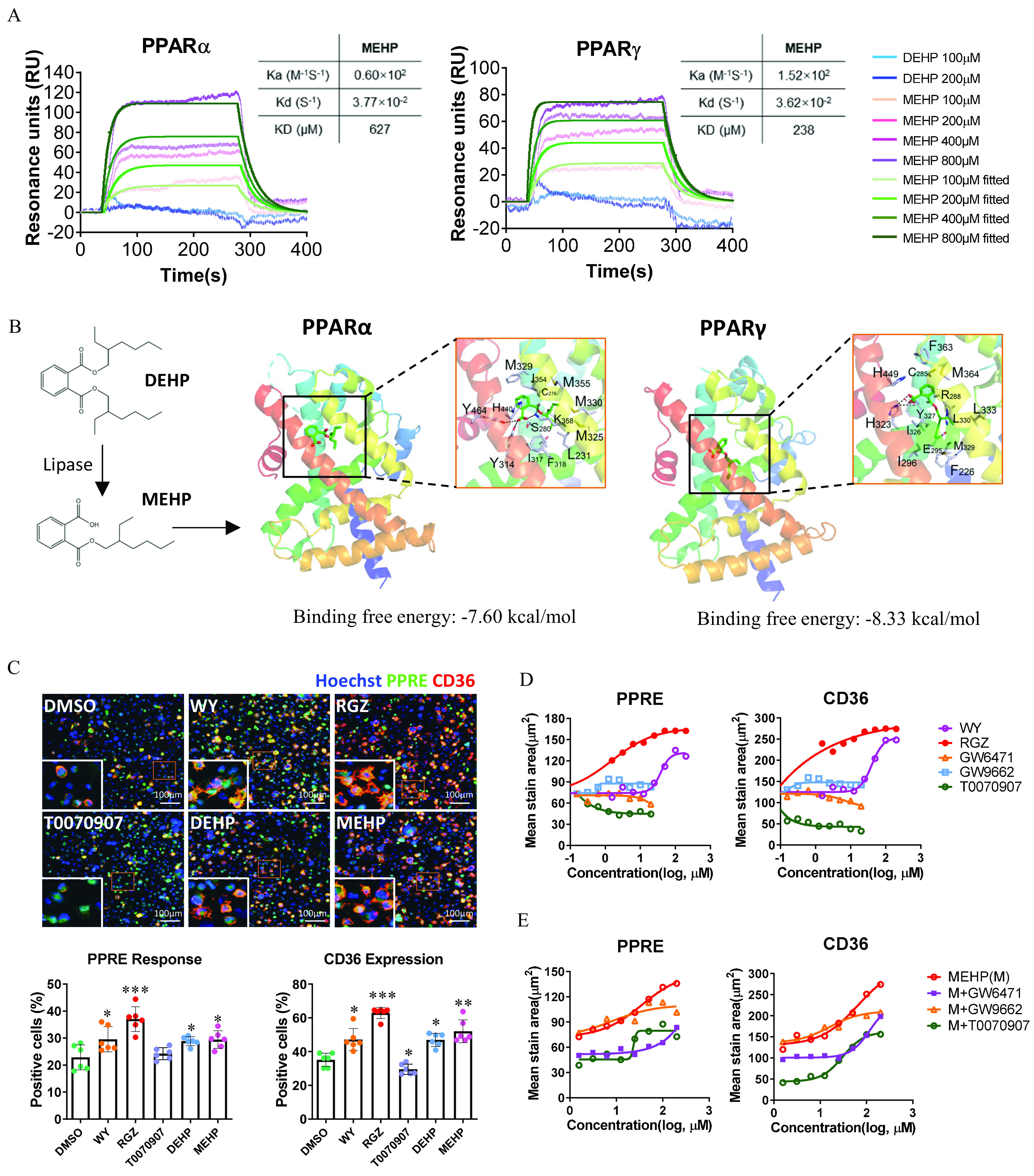
Determination of human and activation by DEHP and MEHP at the molecular and cellular levels. (A) Sensorgrams are shown of the binding responses of DEHP and MEHP with human or proteins. The binding affinity was determined using surface plasmon resonance (SPR). The data are provided in Tables S8 and S9. (B) Molecular docking simulation for the ligand–protein binding of MEHP with human and . The proteins are displayed as ribbons, and small molecules are displayed as sticks. (C) High-content imaging (up) and quantification (down) of PPRE response and CD36 expression in the PPRE-eGEP-THP-1–derived macrophages. The cells were treated with Wy14643 (WY; ), rosiglitazone (RGZ; ), T0070907 (), DEHP (), and MEHP () for 24 h. The number of cells with positive staining of PPRE activation (green) and CD36 expression (red) were normalized to the total cells (blue, stained with Hoechst 33342) in each of triplicate wells (the plots contain all technical replicates from two imaging sites of each well). Data are expressed as . The data are provided in Table S10. The data were analyzed using one-way ANOVA followed by Dunnett’s multiple comparisons test. *, **, *** compared with the vehicle control (DMSO). (D) Dose–response curves of PPRE activation and CD36 expression for selective PPARs ligands. The PPRE-eGEP-THP-1–derived macrophages were treated with selective PPARs ligands at various concentrations of WY (), RGZ (), GW6471 (), GW9662 (), or T0070907 () in replicate wells () for 24 h. The summary data are provided in Tables S11 and S12. (E) Dose–response curves of PPRE activation and CD36 expression for MEHP. PPRE-eGEP-THP-1–derived macrophages were treated with MEHP (M; ) alone or in combination with GW6471 (), GW9662 (), or T0070907 () in replicate wells () for 24 h. The summary data are provided in Table S13. The fluorescent stain of PPRE-eGFP and CD36 were analyzed using high-content cellular imaging, as described in the “Methods” section. values of concentrations were used. Note: ANOVA, analysis of variance; DEHP, diethylhexyl phthalate; DMSO, dimethyl sulfoxide; MEHP, mono(2-ethylhexyl) phthalate; PPAR, peroxisome proliferator-activated receptor; PPRE, peroxisome proliferator-activated receptor response element; RU, resonance unit; SD, standard deviation.
