Figure 3.
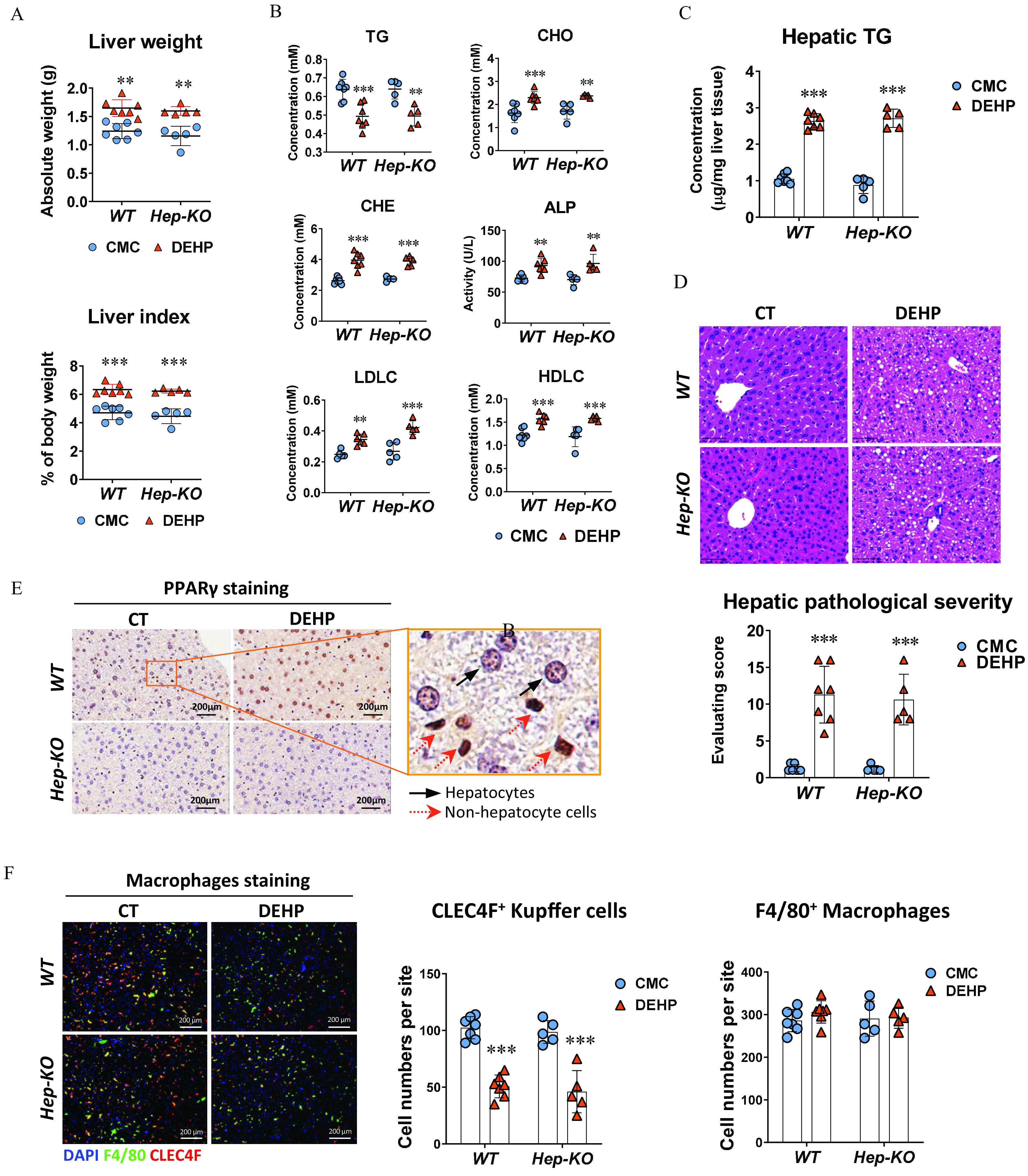
Effect of hepatocyte-specific knockout in DEHP-induced fatty liver. Wild-type (WT; ) and hepatocyte-specific knockout (Hep-KO; ) C57BL/6J male mice were treated with 0.5% (wt/vol) sodium carboxymethylcellulose (CMC; vehicle control) or DEHP ( BW) by daily gavage for 28 d. (A) Liver weight and liver index of mice at the end of the treatment. The data are provided in Table S14. (B) Plasma level of triglycerides (TGs), cholesterol (CHO), high-density lipoprotein cholesterol (HDLC), low-density lipoprotein cholesterol (LDLC), alkaline phosphatase (ALP), and cholinesterase (CHE) of WT () and Hep-KO () mice at the end of the treatment. The data are provided in Table S15. (C) Hepatic TG concentration in the liver of WT () and Hep-KO () mice at the end of the treatment. The data are provided in Table S16. (D) Representative histology images (up) and quantification of hepatic steatosis (down) in the WT () and Hep-KO () mice at the end of the treatment. The data are provided in Table S17. (E) Representative immunohistochemistry (IHC) images of expression in the liver of WT and Hep-KO mice at the end of the treatment. (F) Immunofluorescence staining of CLEC4F (red) and F4/80 (green) in the mouse liver (left). Cell nuclei were stained with DAPI (blue). Cells with positive stain in the liver sections of WT () and Hep-KO () mice were quantified using the ImageXpress system under . The data are provided in Table S18. All data are expressed as . The data were analyzed using one-way ANOVA followed by Bonferroni’s multiple comparisons test. *, **, *** compared with the vehicle control (CMC). Note: ANOVA, analysis of variance; BW, body weight; CT, control; DAPI, 4′,6-diamidino-2-phenylindole; DEHP, diethylhexyl phthalate; Hep-KO, hepatic PPAR knockout mice; Mac-KO, macrophage-specific PPAR knockout mice; PPAR, peroxisome proliferator-activated receptor; SD, standard deviation.
