ABSTRACT
In 2017, the Iowa State University Veterinary Diagnostic Laboratory detected a reverse-zoonotic transmission of a human seasonal H3 influenza A virus into swine (IAV-S) in Oklahoma. Pairwise comparison between the recently characterized human seasonal H3 IAV-S (H3.2010.2) hemagglutinin (HA) sequences detected in swine and the most similar 2016–2017 human seasonal H3 revealed 99.9% nucleotide identity. To elucidate the origin of H3.2010.2 IAV-S, 45 HA and 27 neuraminidase (NA) sequences from 2017 to 2020 as well as 11 whole-genome sequences (WGS) were genetically characterized. Time to most recent common human ancestor was estimated between August and September 2016. The N2 NA was of human origin in all but one strain from diagnostic submissions with NA sequences, and the internal gene segments from WGS consisted of matrix genes originating from the 2009 pandemic H1N1 and another 5 internal genes of triple reassortant swine origin (TTTTPT). Pigs experimentally infected with H3.2010.2 demonstrated efficient nasal shedding and replication in the lungs, mild pneumonia, and minimal microscopic lung lesions and transmitted the virus to indirect contact swine. Antigenically, H3.2010.2 viruses were closer to a human seasonal vaccine strain, A/Hong Kong/4801/2014, than to the H3.2010.1 human seasonal H3 viruses detected in swine in 2012. This was the second sustained transmission of a human seasonal IAV into swine from the 2010 decade after H3.2010.1. Monitoring the spillover and detection of novel IAV from humans to swine may help vaccine antigen selection and could impact pandemic preparedness.
IMPORTANCE H3.2010.2 is a new phylogenetic clade of H3N2 circulating in swine that became established after the spillover of a human seasonal H3N2 from the 2016–2017 influenza season. The novel H3.2010.2 transmitted and adapted to the swine host and demonstrated reassortment with internal genes from strains endemic to pigs, but it maintained human-like HA and NA. It is genetically and antigenically distinct from the H3.2010.1 H3N2 introduced earlier in the 2010 decade. Human seasonal IAV spillovers into swine become established in the population through adaptation and sustained transmission and contribute to the genetic and antigenic diversity of IAV circulating in swine. Continued IAV surveillance is necessary to detect emergence of novel strains in swine and assist with vaccine antigen selection to improve the ability to prevent respiratory disease in swine as well as the risk of zoonotic transmission.
KEYWORDS: influenza A virus, H3N2, human-to-swine spillover, reverse zoonosis, swine, H3.2010.2
INTRODUCTION
Influenza A virus (IAV) is a rapidly evolving, segmented, RNA virus endemic to swine (IAV-S) that undergoes genetic change due to polymerase error and reassortment, and it poses a threat to both human and animal health (1–3). Epithelial cells lining the swine respiratory tract express α2,6- and α2,3-Gal-linked sialic acid on their cell surface that may make them vulnerable to IAV infections from other species. Swine infected with human- or avian-origin IAV can generate viruses with unique combinations of surface and internal genes, allowing pigs to be potential mixing vessels for the generation of novel viruses that may contribute to IAV genetic diversity (4).
Influenza viruses capable of interspecies transmission have caused pandemics in the past, such as the 1918 Spanish flu, which later became the origin of the classical H1 lineage in swine (5). The 1990 decade marked the spillover of human seasonal H3N2 IAV into North American swine generating novel viruses with the triple reassortant internal gene constellation (TRIG), which caused a dramatic change in the genetic ecology of IAV circulating in swine (6). Since the 2009 H1N1 pandemic (H1pdm09), increased surveillance efforts have identified additional spillover events where H1 and H3 viruses transmitted from humans to swine have become established in the pig population and impacted the genetic diversity of IAV in swine globally (6–10). The transmission of human seasonal H3N2 and pre-2009 H1N1 viruses into swine have been followed by adaptation that includes changes in glycosylation sites in the hemagglutinin (HA) gene, amino acid substitutions, and reassortment with IAV endemic to pigs (11). In contrast, H1pdm09 transmitted from human to swine more frequently maintains the original internal gene segments of H1pdm09, with limited adaptive changes required for infection and potential sustained transmission in swine, and has been involved in yearly human-to-swine transmission and outbreaks of clinical disease in pigs (8, 10, 12, 13). These events highlight the need to monitor the evolution of IAV in pigs to better prepare for potential human pandemics that could involve swine-lineage IAV (8).
Currently, three IAV subtypes, H1N1, H1N2, and H3N2, cocirculate in U.S. swine. Although IAV can transmit from swine to humans, spillovers of virus from humans to swine is common (3, 10). At least 20 introductions of human seasonal IAV into swine, 13 of them H3N2, were identified between 1965 and 2013, although most did not become endemic (9). The most recent successful human-to-swine spillover with sustained transmission in pigs was a human seasonal H3 that was detected in 2011 (13). This IAV was designated the H3.2010.1 clade in swine (14). Since 2016, H3.2010.1 became a predominant H3 genetic clade detected at the Iowa State University Veterinary Diagnostic Laboratory (ISU VDL) and in 2020 accounted for approximately 39% of the H3 HA sequences (15). In 2020, six H3 phylogenetic clades, H3.1990.4 (cluster IV, or C-IV), H3.1990.4A (C-IVA), H3.1990.4B (C-IVB), H3.2010.1, H3.1990.1 (cluster I, or C-I), and the newly designated H3.2010.2, described here, cocirculated in U.S. swine (15).
H3.2010.2, the most recent human-to-swine H3 spillover with sustained transmission in the United States, was detected in 2016 and is the second human seasonal H3 to become established in swine in that decade. The H3.2010.2 clade was detected in approximately 3.4% of H3 IAV sequences generated at the ISU VDL from 2017 to 2020 (15). The aim of the current study was to genetically and antigenically characterize the novel H3.2010.2 and evaluate its pathogenesis and transmission in swine.
RESULT
Case history.
From January 2017 to December 2020, 45/1,362 H3 HA sequences were identified that did not belong to known phylogenetic clades circulating in U.S. swine, although their consistent detection suggested sustained transmission in swine (16). These 45 uncharacterized H3 HA from the ISU VDL were subjected to NCBI BLAST, which revealed these HA were 99.9% similar to the closest matching H3 HA from the 2016–2017 human influenza season (A/Indiana/23/2017; GenBank accession no. CY234634.1). Spatial dissemination of the 45 ISU VDL human-like H3 HA across the central U.S. included 17 from Indiana, 10 from Oklahoma, 9 from Iowa, 5 from Illinois, and 4 from Ohio. Due to the continuation of detections over multiple years and evidence of regional spread, the HA genes were designated H3.2010.2 as the second sustained H3 introduction of the 2010 decade.
The 45 H3.2010.2 sequences were associated with accessions submitted to the ISU VDL for diagnostic purposes that represented at least 6 different production systems and 19 sites originating from the 5 states. There were 13 accessions with H3.2010.2 HA sequences detected exclusively through ISU VDL submissions to the anonymous USDA IAV surveillance system in swine that provides only the date of collection, specimen type, and state of detection. The pig age or stage of production was represented by 8 accessions from breeding/farrowing operations, 2 from replacement stock, 14 from nursery, and 6 from grow-finish, and 2 accessions did not specify a stage of production on the submission form. Three accessions demonstrated mixed infections with H1 IAV, and one accession detected an H3 HA with an N1 and N2 NA.
Clinical epidemiology data.
Among the 45 ISU VDL accessions containing H3.2010.2 HA sequences, 17 accessions were submitted for routine monitoring and requested only quantitative reverse transcriptase real-time PCR (qRT-PCR) screening, subtyping, and HA sequencing without providing clinical epidemiology data beyond the production system, site, state, sample type, and stage of production. Clinical signs of respiratory disease (coughing) were reported in 11 accessions in addition to variable reports of lethargy and wasting. Pneumonic lesions were reported by the submitter in 8 accessions and included lung consolidation, suggestive of influenza, and 1 of the 8 accessions also reported pleuritis, suggesting secondary bacteria. There were 8 accessions reporting necrotizing bronchiolitis consistent with IAV-S microscopic lesions. Some coinfections were identified by qRT-PCR, including 2 accessions that detected porcine reproductive and respiratory syndrome virus (PRRSV) and 1 accession that detected Mycoplasma hyorhinis by qPCR. Routine culture detected secondary bacteria in 10 accessions, including single or coinfections of Streptococcus suis, Glaesserella parasuis, Actinobacillus suis, Pasteurella multocida type D, and Bordetella bronchiseptica.
Genetic characterization.
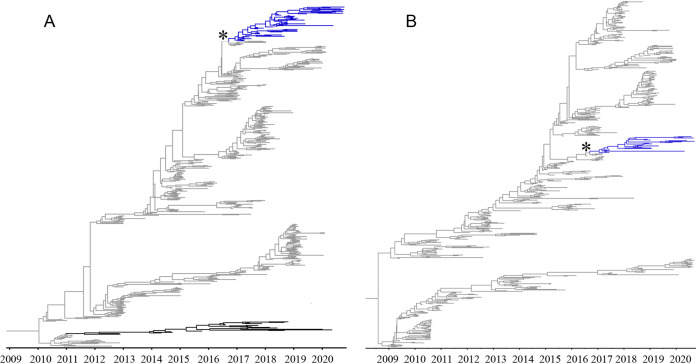
A maximum likelihood (ML) analysis was conducted with the 45 ISU VDL H3.2010.2 HA sequences (see Table S1 in the supplemental material) and the following HA sequences that were randomly sampled from the Influenza Research Database (IRD): 10 H3.2010.2, 35 cluster I (C-I), 248 C-IV (11 C-IV, 199 C-IVA, 37 C-IVB, and 1 C-IVE), and 575 H3.2010.1 (Fig. S1A). For the NA maximum likelihood phylogenetic analysis, 27 ISU VDL N2 NA and 33 2016, 22 1998, 6 1998B, 209 2002A, and 579 2002B from the IRD were used (Fig. S1B). The 55 IAV H3.2010.2 HA sequences clustered together in a monophyletic clade with a common ancestor from the human seasonal H3 HA sequences (Fig. S1A). Time-scaled Bayesian phylogenetic trees of the H3.2010.2 HA and NA genes were reconstructed for swine to estimate the time to the most recent human seasonal H3 ancestor (Fig. 1). All 55 H3.2010.2 HA sequences shared a common ancestor with human seasonal H3 IAV and formed a separate, statistically supported clade (Fig. 1A). The maximum clade credibility phylogenetic tree revealed the time of the H3.2010.2 spillover from humans to swine was approximately August to September 2016 (2016.53 to 2016.85, 95% highest posterior density [HPD]). The current phylogenetic tree reflects only one independent human-to-swine H3N2 introduction of H3.2010.2 without evidence of additional spillover events. The HA nucleotide diversity within the H3.2010.2 clade averaged 1%, and between-clade divergences were 3% with human seasonal strains, 7% with H3.2010.1 in swine, and up to 15% with swine C-IV, C-IVA, and C-IVB (Table 1). The within- and between-clade percent identity warranted the 2010.2 clade designation for swine H3 nomenclature. Average amino acid differences between the H3.2010.1 and H3.2010.2 HA chosen for antigenic analysis were 11% (Table S2 to S4).
FIG 1.
Time scaled phylogenies demonstrating time of spillover of human seasonal H3N2 virus into swine. (A) Bayesian phylogenetic tree of the H3.2010.2 HA, including human seasonal H3 HA sequences. (B) Bayesian phylogenetic tree of the H3.2010.2 N2 NA, including human seasonal N2 NA sequences. Human H3-HA and N2-NA data are colored gray, swine data (H3.2010.1) are colored black, and H3. 2010.2 data are colored blue. The asterisk indicates the spillover event from humans to swine.
TABLE 1.
Estimates of evolutionary divergence over sequence pairs of swine and human H3 HA within and between phylogenetic cladesa
| H3 clade | Within clade (%) | Between clades (%) |
||||
|---|---|---|---|---|---|---|
| C-IV | C-IVA | C-IVB | 3.2010.1 | 3.2010.2 | ||
| C-IV | 8.3 (2.0–12.0) | |||||
| C-IVA | 3.0 (0.0–7.0) | 9.6 (8.4–12.0) | ||||
| C-IVB | 5.2 (0.3–10.1) | 9.8 (9.0–12.0) | 9.0 (8.0–11.0) | |||
| 3.2010.1 | 2.0 (0.0–5.0) | 15.0 (14.0–17.0) | 14.4 (13.0–17.0) | 14.7 (13.0–17.0) | ||
| 3.2010.2 | 0.9 (0.0–2.3) | 15.0 (14.0–16.0) | 14.6 (13.5–16.0) | 14.9 (14.0–16.0) | 7.1 (6.0–9.0) | |
| Hu seasonalb | 2.0 (0.0–4.0) | 14.6 (13.0–16.0) | 14.4 (13.0–16.0) | 14.4 (13.0–16.0) | 6.6 (5.0–8.0) | 2.5 (0.2–5.0) |
Ranges of divergence between sequences are shown in parentheses.
Hu seasonal, human seasonal IAV from years 2016 to 2020.
Maximum likelihood (ML) phylogenetic trees. (A) ML phylogenetic tree of the swine H3-HA clades currently circulating in swine. The novel H3.2010.2 HA clade is highlighted in blue. (B) ML phylogenetic tree of the swine N2-NA clades currently circulating in swine. H3.2010.2 NA clade is highlighted in blue. The branches are colored based on phylogenetic clade designation. Download FIG S1, TIF file, 0.2 MB (162.5KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Swine influenza A virus cases detected in the ISU VDL from the recent sustained human to swine spillover from 2017 to 2020 in the United States. Download Table S1, PDF file, 0.04 MB (41.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Representative H3.2010.1 and H3.2010.2 influenza A virus from swine used in the antigenic distance analysis. Download Table S2, PDF file, 0.01 MB (13.1KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The 11 HA sequences generated from MiSeq whole-genome sequencing (WGS) were consistent with single HA gene Sanger sequencing results. The H3.2010.2 WGS data demonstrated the NA genes were closely related to 2016 human seasonal H3N2, the PB2, PB1, PA, NP, and NS genes originated from the triple reassortant internal gene constellation (TRIG), and the M genes originated from the H1pdm09 lineage (see Fig. S2 to S7 at 10.6084/m9.figshare.16934590). The time of the 2016 human seasonal NA introduction in swine matched the H3 HA, which was approximately August to September 2016 (2016.53 to 2016.85, 95% HPD) (Fig. 1B). Phylogenies inferred for individual internal gene segments (see Fig. S2 to S7 at 10.6084/m9.figshare.16934590) suggested reassortment of the human seasonal H3N2 with IAV endemic to swine where the virus acquired HA and NA from human seasonal IAV and internal genes from IAV-S. One WGS was identified to have acquired N2-NA of 2002B swine lineage. Phylogenies suggest a single introduction of the virus followed by reassortment events.
Antigenic characterization and glycosylation pattern.
Six amino acid positions (145, 155, 156, 158, 159, and 189; H3 numbering) in the globular head region of the hemagglutinin gene (HA1 domain) are known to play an important role in defining the antigenic phenotype of IAV-S (17). These six positions collectively are referred to as the antigenic motif. There were two predominant antigenic motifs, STHNYK and STHNYN, detected in the majority of H3.2010.2 ISU VDL HA sequences from 2017 to 2020. The STHNYK motif was predominant in H3.2010.2 in 2017, which was also the antigenic motif identified in 2016–2017 human seasonal H3 sequences, and most of the H3.2010.2 sequences demonstrating the STHNYK antigenic motif detected in 2017 were from Oklahoma. Subsequently, an STHNYN motif became predominant after 2017.
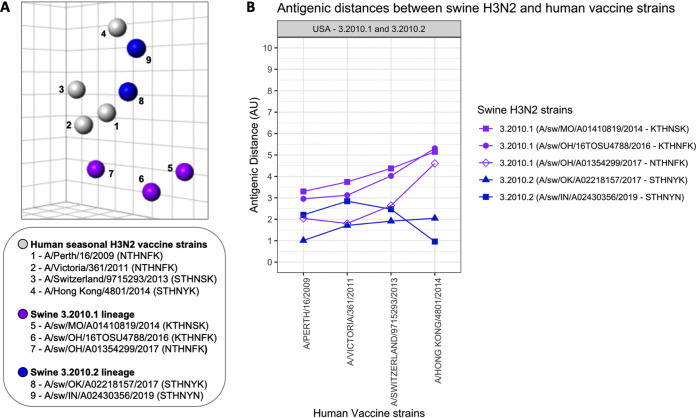
An antigenic map was generated to visualize antigenic relationships between representative swine H3N2 strains of the H3.2010.2 lineage, the H3.2010.1 lineage (Table S2), and human seasonal H3N2 vaccine strains using hemagglutination inhibition (HI) data (Fig. 2A) (Table S5). The H3.2010.2 swine viruses retained closer antigenic similarity to human seasonal H3N2 than the H3.2010.1 swine viruses. A/swine/Oklahoma/A02218157/2017 (OK/17; GenBank accession no. MF471685), representing H3.2010.2 with an STHNYK motif, demonstrated the closest antigenic relationship (1.0 antigen unit [AU]) to the human seasonal H3N2 vaccine strain A/Perth/16/2009, whereas A/swine/Indiana/A02430356/2019 (IN/19; GenBank accession no. MK640458), representing H3.2010.2 with an STHNYN motif, demonstrated the closest antigenic relationship (1.0 AU) to a genetically similar human seasonal H3N2 vaccine strain, A/Hong Kong/4801/2014 (Fig. 2B, Table S5).
FIG 2.
Antigenic relationships between swine H3.2010.1 and H3.2010.2 lineages and human seasonal H3N2 vaccine strains. (A) Three-dimensional antigenic map of H3.2010 lineages and human seasonal H3N2 vaccine strains. Spheres represent each antigen in the map colored by lineage: H3.2010.1 (purple), H3.2010.2 (blue), and human seasonal H3N2 vaccine strains (gray). Numbers in the map indicate strain name with antigenic motif in parentheses in the legend. Each square of the background grid in the map corresponds to 1 antigenic unit (AU), which represents a 2-fold loss in HI cross-reactivity. (B) Antigenic distances of swine H3.2010.1 and H3.2010.2 lineages from recent human seasonal H3N2 vaccine strains. The antigenic distance is reported in antigenic units (AU).
HA1 percentage of amino acid identity of H3.2010.1 and H3.2010.2 strains used in the antigenic distance analysis. Download Table S4, PDF file, 0.01 MB (13.9KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Cross hemagglutination inhibition (HI) titers representing influenza A viruses from the antigenic map and antigenic distance analysis. Download Table S5, PDF file, 0.02 MB (22.1KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
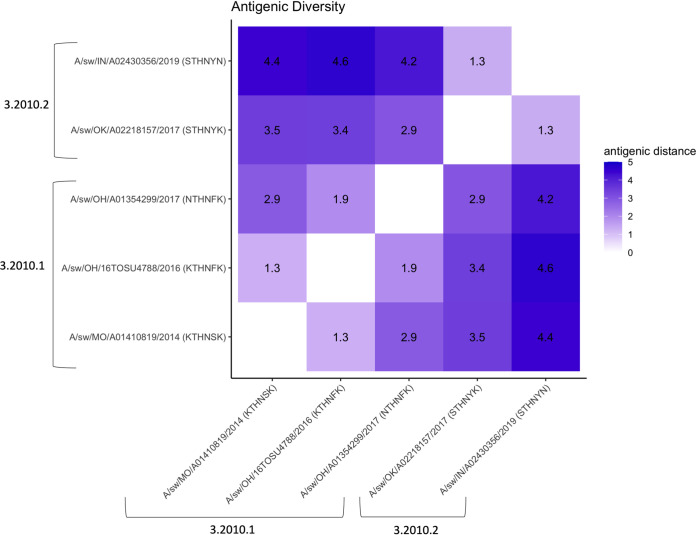
A heat map with antigenic distances was generated to demonstrate the antigenic diversity between H3.2010.1 and H3.2010.2 swine lineages (Fig. 3). The representative strains within the H3.2010.2 lineage, OK/17 (STHNYK) and IN/19 (STHNYN), were antigenically similar, within 1.3 AU from each other. In contrast, both H3.2010.2 strains were at minimum 2.9 AU away from representative strains of the H3.2010.1 lineage (Table S3), indicating the swine strains represented from clade H3.2010.2 formed a separate antigenic cluster from H3.2010.1. Fourteen amino acid positions were predicted as putative N-glycosylated sites in the initial H3.2010.2 clade HA peptides. From late 2017 through 2020, there was a complete loss of one putative N-glycosylation site at amino acid position 133 (H3 numbering).
FIG 3.
Heat map of antigenic diversity based on the pairwise antigenic distances between swine H3.2010.1 and H3.2010.2 lineages. The antigenic distances are in antigenic units (AU), where 1 AU represents a 2-fold loss in HI cross-reactivity.
Hemagglutinin (HA) amino acid sequence alignment of the H3.2010.1 and H3.2010.2 swine lineages used for antigenic analysis. Identity to the consensus sequences is represented by a dot. Antigenic motif (H3 numbering: 145, 155, 156, 158, 159, 189) is highlighted with red boxes. Download Table S3, PDF file, 0.2 MB (186.9KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Pathogenesis and transmission study.
Pigs were challenged with IN/19, representing the H3.2010.2 swine strains from 2018 to 2019, to evaluate pathogenesis and transmission. At 5 days postinfection (dpi), challenged pigs demonstrated mild macroscopic lung lesions typical of IAV infection, with a group average of 6% lung consolidation (Table 2). The magnitude of microscopic lung lesion scores (average score, 4.6) were consistent with the macroscopic lesions (Table 2), and microscopic trachea lesion scores were consistent with IAV infection (average score, 2.6) (Table 2). The IN/19 strain replicated at high titers in the lungs (average, 6.2 log10 50% tissue culture infectious dose [TCID50/ml]) in inoculated pigs, and virus shedding was detected in nasal swabs from 1 to 5 dpi (Table 3). Indirect virus transmission to contact pigs was detected by 5 days postcontact (dpc), albeit with low nasal titers, and was confirmed by seroconversion of all contact pigs at 14 dpc (Table 4).
TABLE 2.
Macroscopic and microscopic lung lesions from pigs challenged with H3.2010.2 A/swine/Indiana/A02430356/2019a
| Group | Lung lesions |
Trachea lesions |
|
|---|---|---|---|
| Macroscopic (%) | Microscopic (0–22) | Microscopic (0–8) | |
| Negative control | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| IN/19 | 6.0 ± 1.3b | 4.7 ± 0.6b | 2.7 ± 0.3b |
Values are means ± standard errors of the means (SEM). Microscopic lung lesion scores ranged from 0 to 22, and microscopic trachea lesion scores ranged from 0 to 8.
Significantly different from negative control (P ≤ 0.05).
TABLE 3.
Virus titers in pigs challenged with H3.2010.2 IN/19
| Group | Nasal swab virus titers atb: |
BALF titer at 5 dpi | |||||
|---|---|---|---|---|---|---|---|
| 0 dpi | 1 dpi | 2 dpi | 3 dpi | 4 dpi | 5 dpi | ||
| NCa | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| IN/19 | 0.0 ± 0.0 (0/0) | 2.0 ± 0.4* (7/10) | 2.6 ± 0.5* (8/10) | 2.6 ± 0.3* (9/10) | 3.7 ± 0.3* (10/10) | 3.6 ± 0.2* (10/10) | 6.2 ± 0.1* (10/10) |
NC, nonchallenged controls.
Virus titers are represented as log10 TCID50/ml. Numbers in parentheses indicate number of positive pigs out of the total number of pigs. Data are means ± SEM. An asterisk indicates significantly different from the NC (P ≤ 0.05).
TABLE 4.
Virus titers in pigs that were indirect contacts of pigs infected with H3.2010.2 IN/19
| Group | Nasal swab virus titers ata: |
HI titer at 14 dpcb | ||||||
|---|---|---|---|---|---|---|---|---|
| 1 dpc | 2 dpc | 3 dpc | 4 dpc | 5 dpc | 7dpc | 9dpc | ||
| IN/19 | 0.0 ± 0.0 (0/0) | 0.0 ± 0.0 (0/0) | 0.0 ± 0.0 (0/0) | 0.0 ± 0.0 (0/0) | 0.5 ± 0.5 (1/5) | 1.4 ± 0.9 (2/5) | 1.4 ± 0.9 (2/5) | 208 ± 48 (5/5) |
Virus titers are represented as log10 TCID50/ml. Numbers in parentheses indicate number of positive pigs out of the total number of pigs. Data are means ± SEM.
Geometric mean titer.
DISCUSSION
In the current study, we described the successful incursion and sustained transmission of a second human seasonal H3 IAV into the U.S. swine population in the 2010 decade following the introduction of the H3.2010.1 HA clade. Pairwise comparison between the initial detection of an H3.2010.2 HA sequence in swine and the most similar human seasonal H3 revealed >99% nucleotide identity. The HA sequences of the newly emerged H3.2010.2 first detected in March of 2017 were genetically most similar to H3N2 strains from the 2016–2017 human influenza season, indicating this human H3 IAV was detected relatively soon after its introduction into swine. The H3.2010.2 strains also demonstrated an antigenic phenotype similar to that of recent human seasonal H3 strains, including the A/Hong Kong/4801/2014 human H3 vaccine strain.
Several human seasonal IAV spillovers have occurred in swine that established and became frequently detected throughout North America (18, 19). The first H3N2 was identified in the U.S. swine population in 1998 with an HA from Fujian‐like human seasonal IAV (10, 20). Such transmissions were followed by evidence of reassortment with lineages endemic to swine. Based on phylogenetic analysis, these human seasonal H3N2 variants later evolved into H3 C-IV, which continues to circulate in swine in North America (21, 22). A more recent human seasonal H3 spillover that is currently prevalent in U.S. swine was detected in 2012 and designated the H3.2010.1 clade (10, 13). H3.2010.1 is another example of a successful reverse zoonotic transmission from humans to swine that demonstrated limited detections for several years after the introduction in 2010 to 2011 but accounted for approximately 38% of the ISU VDL H3 HA sequences detected in swine in 2020. These spillover events and other sporadic transmission of human seasonal IAV into swine, followed by adaptation and reassortment, increased the antigenic diversity of IAV circulating in U.S. swine.
Our in vivo study demonstrated that H3.2010.2 (IN/19 strain) with a genome constellation of TTTTPT and human-origin N2-2016 replicated efficiently in the nose and lungs, causing a typical IAV infection in pigs with mild pneumonia. The IN/19 strain transmitted indirectly to contact pigs, similar to previous observations with the H3.2010.1 and other swine-adapted strains (10). The IAV surface glycoproteins (HA and NA) and combination with the internal gene constellation can affect virulence and efficiency of virus replication (23–25). Different combinations of swine H3N2 genome constellations have shown variable degrees of pathology and distinct patterns of viral shedding and transmission in pigs (26). The internal gene constellation plays an important role in the evolution and diversity of the human-origin viruses in swine (26). All 11 of the H3.2010.2 WGS generated in this study contained a TTTTPT internal gene constellation. The M gene from H1pdm09 became predominant in IAV circulating in North American swine after approximately 2014, and the TTTTPT internal gene constellation remains one of the most frequently detected patterns in U.S. swine IAV WGS data. Circulation of such reassortant viruses not only affects the pig industry but also could have public health implications.
Human-to-swine spillover IAV that are maintained in swine are often followed by reassortment of the internal genes with swine-lineage segments, suggesting that reassortment and swine adaptation are important for sustained onward transmission (27). In addition, the transmission of human seasonal IAV to swine and reassortment was also associated with antigenic evolution in the H3.2010.1 swine IAV lineage, as shown here and in previous studies (12, 13). In the current study, the representative H3.2010.2 IAV from 2017 and 2019 were antigenically distinct from H3.2010.1 IAV currently circulating in the U.S. and demonstrated a close antigenic relationship with human seasonal H3N2 vaccine strains. However, human seasonal H3N2 vaccine strains were not available after 2014 in our panel to assess antigenic drift from more current human seasonal H3N2 vaccine strains (Fig. 2).
Substitutions in 7 amino acid positions (145, 155, 156, 158, 159, 189, and 193) were shown to be largely responsible for the antigenic evolution of H3N2 viruses circulating in humans for 35 years (28) and in swine H3N2 strains (17). The antigenic motifs detected in H3.2010.2 in this study were not observed in the 20 most frequently detected cluster-IV H3N2 antigenic motifs in the U.S. from 2012 to 2016 (22). The predominant HA antigenic motif of the initial H3.2010.2 detected in 2017 was STHNYK, similar to contemporary 2016 human seasonal H3 HA, but mutated to STHNYN in the 2018–2020 HA sequences. This change of K to N at position 189 may be the result of adaptation of the virus from humans to the swine host or due to immune pressure in the swine population. Previous reports tested the antigenic effect of substitutions introduced into the motif near the receptor binding site of the HA using an isogenic swine lineage virus (17, 22). It was reported that amino acid substitutions at positions 145 and 189 significantly affected the antigenic phenotype of the virus by substantial loss in HI antibody cross-reactivity. Our antigenic analysis demonstrated the two representative H3.2010.2 strains (STHNYK and STHNYN motifs) were antigenically similar but were 1.3 AU apart and showed a trend for differences in cross-reactivity with the other test antigens that may be explained by the amino acid difference in the motif at position 189 (Fig. 3). Amino acid substitutions outside the motif can also contribute to variations in antigenic phenotype of strains with similar motifs or intracluster antigenic drift (22). Based on antigenic motif, phenotypic differences were observed among cocirculating C-IV IAV in swine (22), but it is unclear if the antigenic evolution of H3.2010.2 HA will follow patterns similar to those of C‐IV or H3.2010.1. The antigenic diversity between H3.2010.1 and H3.2010.2 clades was substantially different, in accordance with changes at two previously described amino acid positions, 145 and 189, that influenced their antigenic properties (28). The antigenic diversity between H3.2010.1 and H3.2010.2 clades was likely also influenced by many other amino acid differences observed outside the antigenic motif (17, 29).
Adaptive changes of H3.2010.2 were suggested by evolution of the antigenic motif and glycosylation pattern over time. Glycosylation of the HA globular head domain is reported to physically shield antigenic sites preventing antibody recognition and leading to viral evasion from antibody-mediated immune responses. Conversely, loss of a glycan may expose a previously shielded antigenic region against which individuals with antibody might be protected (30), but these glycan shields may not be necessary in interspecies transmission to a new naive host, such as 2010.2 H3 introduced from humans to swine. In this study, based on predicted N-glycosylation sites in the translated protein sequences (https://services.healthtech.dtu.dk/service.php?NetNGlyc-1.0), a complete loss of glycosylation was observed at predicted site 133, and a partial loss was also observed at another predicted site, 165, from late 2017 to 2020, although their significance is unknown.
The ISU VDL data suggest that although the spread of H3.2010.2 is limited and may not displace the C‐IV and H3.2010.1 clades that are endemic to the U.S., all three H3 IAV clades continue to cocirculate in U.S. swine. The antigenic differences between H3.2010.1 and H3.2010.2 lineages have relevant implications for vaccine effectiveness and IAV control strategies in U.S. swine populations. In addition, human seasonal H3 IAV transmitted to swine increase the genetic diversity of the virus and complicate the ability of veterinarians to develop effective vaccination strategies. Public health officials also recognize human-like IAV that have become established in swine maintain the potential to cause zoonotic infections. Indeed, the H3.2010.1 HA lineage was responsible for the majority of variant human cases of influenza after circulating in swine for approximately 6 years (31, 32). Rapid detection of the newly emerging virus strains and efficient monitoring of such reassorted viruses and their evolution will help control transmission of the virus in swine and may help with pandemic preparedness.
MATERIALS AND METHODS
Ethics statement.
All cases were submitted to the ISU VDL for general diagnostic purposes, and no ethical compliance for sample collection was required.
IAV diagnostic submissions.
In 2017, two novel human-like H3N2 IAV strains were isolated from swine nasal swab samples submitted to the ISU VDL in the United States (54). The ISU VDL laboratory information management system (LIMS) storing IAV diagnostic test results and associated nucleotide sequences was later searched for H3 sequences from swine and analyzed to determine H3 clade. A total of 45 ISU VDL accessions contained H3 HA sequences from LIMS that did not cluster with previously designated H3 phylogenetic clades and formed a separate monophyletic clade. ISU VDL submissions that were qRT-PCR positive with threshold cycle (CT) values of ≤38 were subtyped using a specific qRT-PCR.
IAV real-time RT-PCR.
IAV extraction was performed on lung, nasal swab, and oral fluid samples submitted to the ISU VDL from 45 accessions. Viral RNA was extracted using the MagMAX pathogen RNA/DNA isolation kit (Thermo Fisher Scientific, Waltham, MA, USA) and a Kingfisher96 instrument (Thermo Fisher Scientific) using the high-volume lysis (HVL) procedure per the manufacturer’s instructions. For the lysis step, 100 μl of sample was added to 240 μl of lysis/binding solution. Xeno internal control RNA was added to the lysis-binding solution at 20,000 copies per reaction prior to extraction to monitor PCR amplification and detect inhibition. The HVL used two washes each of 300 μl and 450 μl wash solutions I and II, respectively. Nucleic acid was eluted in 90 μl elution buffer. The HVL extraction was conducted using the Kingfisher program AM1836_DW_HV_v3.
IAV screening qRT-PCR was performed on nucleic acid extracts according to the manufacturer’s instructions using PCR reagents with multiple primers and probes targeting different genomic regions (VetMAX gold swine influenza virus [SIV] detection kit; Thermo Fisher Scientific). One positive extraction control, one positive amplification control, one negative extraction control, and a negative amplification control were included with each extraction and/or PCR run. Each qRT-PCR included 12.5 μl of 2× multiplex RT-PCR buffer, 1.0 μl of 25× SIV primer probe mix, 2.5 μl of 10× multiplex RT-PCR enzyme mix, and 1.0 μl of nuclease-free water. A final volume of 25 μl, consisting of 17 μl master mix and 8 μl of RNA extract, was placed in each well of a 96-well fast PCR plate (Thermo Fisher Scientific). The qRT-PCR was performed using standard mode on an AB 7500 Fast thermocycler: 1 cycle at 48°C for 10 min, 1 cycle at 95°C for 10 min, and 40 cycles of 95°C for 15 s and 60°C for 45 s. Amplification curves were analyzed with commercial thermal cycler system software. Run data were analyzed using auto baseline with thresholds of the target and Xeno set according to the kit insert. Samples with CT values of <38 were considered positive.
IAV subtyping real-time RT-PCR.
HA and NA subtyping was performed on qRT-PCR-positive IAV nucleic acid extracts using the VetMAX-gold SIV subtyping kit (Thermo Fisher Scientific). Separate qRT-PCRs were used to detect the presence of H1 or H3 HA and N1 or N2 NA, respectively. Each qRT-PCR included 12.5 μl of 2× multiplex RT-PCR buffer, 1.0 μl of 25× H1H3 or N1N2 primer-probe mix, 2.5 μl of 10× multiplex RT-PCR enzyme mix, and 1.0 μl of nuclease-free water. Each subtyping plate included the same positive and negative controls used in the swine IAV general qRT-PCR. Cycling conditions were the same as those for the IAV screening qRT-PCR, and amplification curves were analyzed with commercial thermal cycler system software using the auto baseline with thresholds set as described in the PCR kit insert. Samples with CT values of <38 were considered positive.
Sanger sequencing IAV HA gene.
Whole-gene segments of the HA were sequenced using Sanger methods for initial sequence analysis. Viral RNA was extracted as described above. Conventional RT-PCR was conducted for each gene segment (primers provided upon request) using the qScript XLT 1-step RT-PCR kit (Quantabio, Beverly, MA). The sequencing RT-PCR setup reaction mix was according to manufacturer’s recommendations, using 200 nM each primer. One positive extraction control (H1 or H3), one negative extraction control, and one negative amplification control were included with the reaction. The RT-PCR was performed using an ABI 2720 thermal cycler: 1 cycle at 48°C for 20 min, 1 cycle at 94°C for 3 min, and 45 cycles of 94°C for 30 s, 55°C for 50 s, and 68°C for 150 s. The final elongation step was 68°C for 7 min. Detection of the RT-PCR product, HA at 1,701 bp, was performed on a QIAxcel (Qiagen, Inc., Germantown, MD, USA) capillary electrophoresis system using a DNA screening cartridge and the AM420 method and purified with ExoSAP-IT PCR cleanup reagent (Affymetrix; Thermo Fisher Scientific) by following the manufacturer’s recommendations. Samples were submitted to the Iowa State University DNA facility (Ames, IA) for sequencing. Lasergene software (DNAStar, Madison, WI) was used to compile sequences.
WGS.
WGS was performed on 11 randomly selected IAV isolates or clinical specimens submitted to ISU VDL for general diagnostic purposes. Viral RNA was extracted using the MagMAX pathogen RNA kit (Thermo Fisher Scientific) and a KingFisher Flex system (Thermo Fisher Scientific). Sequencing libraries were constructed using TruSeq (Illumina, Inc., San Diego, CA). Next-generation sequencing was performed on an Illumina, Inc., MiSeq platform by following standard Illumina protocols at the ISU VDL (34, 35). Approximately 2,000,000 raw sequencing reads per sample were preprocessed using Trimmomatic v0.36. Sequencing was quality checked with FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) (36). Quality-trimmed total reads were mapped against reference sequences downloaded from the NCBI Influenza Sequence Database (ftp://ftp.ncbi.nih.gov/genomes/INFLUENZA/) using BWA-MEM (36). Mapped reads were extracted using SAMtools and used for de novo assembly (37). For each segment, contigs were assembled using ABySS and SPAdes (38, 39). The contigs were manually curated in SeqMan Pro to remove extraneous sequences and trim chimeric contigs, generating a consensus sequence for each segment.
Phylogenetic analysis.
Publicly available human seasonal IAV and swine H3 HA sequences were randomly sampled and obtained from the Influenza Research Database (IRD) (www.fludb.org) (40). For the H3-HA Bayesian analysis, 501 human HA and swine H3-HA (H3.2010.1) sequences were randomly sampled, downloaded, and aligned with 55 H3.2010.2 HA (45 from ISU VDL, 10 from IRD) using default settings in MAFFT v7.409 (41). The best-known maximum likelihood phylogenetic tree was inferred for H3 HA using FastTree v2.1.10, implementing a general time-reversible (GTR) model of nucleotide substitution with gamma-distributed rate variation among sites to generate maximum likelihood phylogenetic trees (see Fig. S1 in the supplemental material) (42). The 26 NA (26/27 N2-NA that were of human origin were retained while one case that had N2-NA from swine 2002B lineage was not used in the Bayesian analysis) were aligned with 498 human N2-NA, and phylogenetic trees were constructed. The 11 internal gene sequences were also aligned as described with randomly selected swine IAV H3 sequences (since the internal genes were not of human origin, human data were excluded from Bayesian analysis for the internal genes), and phylogenetic trees were constructed for all internal genes. A time-scaled Bayesian analysis for each of the 8 gene segments was used to compute the time to most recent common human ancestor (Fig. 1A and B; also see Fig. S2 to S7 at 10.6084/m9.figshare.16934590) (43, 44). The time-scaled Bayesian analysis employed a GTR substitution model with estimated base frequencies and uncorrelated relaxed clock. The phylogenetic trees used a constant GMRF Bayesian Skyride tree prior (44). The analysis was run with 100,000,000 Monte Carlo-Markov chain length with 10,000 echostate to screen. Tracer implemented in BEAST was used to check the analysis for convergence, and 10% of the chain was removed as initial burn-in. Treeannotator was used for summarizing trees created by BEAST 1.10 (44). Figtree v1.4.4 was used to view and annotate the phylogenetic trees (http://tree.bio.ed.ac.uk/software/figtree/).
Within- and between-clade nucleotide distances for the H3 HA clades were calculated in MEGA-X (45). OctoFLU was used for annotation of the internal genes (https://github.com/flu-crew/octoflu) (14). H3 HA glycosylation patterns were analyzed using the NetNGlyc online tool (https://services.healthtech.dtu.dk/service.php?NetNGlyc-1.0). Six H3 HA amino acid positions, 145, 155, 156, 158, 159, and 189 (H3 numbering), found in the globular head of the HA gene, are important to antigenic evolution in swine. These positions were collectively referred to as the antigenic motif and are known to play a role in defining the antigenic phenotype (28, 46). These positions were identified in the sequences based on alignment and using H3 numbering. Microreact (http://microreact.org) was used for spatial distribution data visualization. Data used in the study can be accessed through the following link: https://microreact.org/project/4AiXTceagFHQjH7uoZrhUH.
Antigenic characterization.
Monovalent antisera were raised in pigs against human seasonal H3N2 vaccine strains (A/Perth/16/2009, A/Victoria/361/2011, A/Switzerland/9715293/2013, and A/Hong Kong/4801/2014) and representative swine strains of H3.2010.1 and H3.2010.2 lineages (see Table S2 in the supplemental material). Prior to immunization, all pigs were treated with ceftiofur crystalline-free acid (Excede; Pfizer, New York, NY) and enrofloxacin (Baytril; Bayer Animal Health, Shawnee Mission, KS). A pair of 3-week-old naive pigs were immunized with two doses of IAV whole inactivated vaccine (WIV) adjuvanted with Emulsigen-D (MVP Laboratories, Inc., Ralston, NE) 2 weeks apart. Blood was collected when pigs reached an HI titer of ≥160, as previously described (47). Pigs were cared for in compliance with the Institutional Animal Care and Use Committee of the National Animal Disease Center, USDA-ARS. Sera were treated with a receptor-destroying enzyme (RDE) (Sigma-Aldrich, MO, USA), heat inactivated at 56°C for 30 min, and adsorbed with 50% turkey red blood cells to remove nonspecific agglutination and hemagglutination inhibitors, and HI assays were performed according to a previous protocol (48). Cross-HI data were used to quantify antigenic distances using antigenic cartography as previously described, in which 1 antigenic unit (AU) is equivalent to a 2-fold loss in HI cross-reactivity (17). Antigenic distances were plotted using ggplot2 in R Studio version 1.1.383 (49, 50).
In vivo pathogenesis and transmission study.
Pigs were obtained from a herd free of IAV and porcine reproductive and respiratory syndrome virus (PRRSV). Upon arrival, pigs were treated prophylactically with ceftiofur (Zoetis, Florham Park, NJ), according to the label directions, to reduce potential respiratory bacterial pathogens. Pigs were housed in biosafety level 2 (BSL2) containment during the challenge. Pigs were cared for in compliance with the Institutional Animal Care and Use Committee of the National Animal Disease Center, USDA-ARS, Ames, Iowa. Swine IAV strains were grown in Madin-Darby canine kidney (MDCK) cells with Opti-MEM (Life Technologies, Carlsbad, CA), and clarified virus from infected culture was used for challenge. Ten 3-week-old cross-bred pigs were group-housed in a single isolation room and challenged with 2 ml intratracheally and 1 ml intranasally 105 TCID50/ml A/swine/Indiana/A02430356 /2019(H3N2), and 5 noninfected pigs housed in a separate isolation room served as negative controls. To assess virus transmission, at 2 dpi 5 naive pigs were placed in the same room as IAV-inoculated pigs but in a different pen as indirect contacts. Nasal swabs were collected daily from primary pigs from 0 through 5 dpi and from contact pigs from 0 through 5 dpc and then on 7 and 9 dpc. Ten primary inoculated pigs and five negative-control pigs were humanely euthanized and necropsied at 5 dpi to evaluate lung pathology and virus replication. Bronchoalveolar lavage fluid (BALF) samples of all primary pigs were collected at necropsy (5 dpi). Serum samples were collected from contact pigs at 14 dpc to assess seroconversion and confirm transmission at the time of euthanasia.
Pathology.
The percentage of the lung affected with purple-red consolidation typical of IAV in swine was visually estimated at 5 dpi as previously described (51). Tissue samples from the trachea and right middle or affected lung lobe were fixed in 10% buffered formalin for histopathologic examination. Tissues were processed by routine histopathologic procedures, slides stained with hematoxylin and eosin (H&E), and lesions scored by a veterinary pathologist (51).
Virus titration.
Nasal swab and BALF samples were titrated on MDCK cells to evaluate virus replication in the nose and lungs, as previously described (52). MDCK-inoculated monolayers were evaluated for cytopathic effect (CPE) between 48 and 72 h postinfection, fixed with 4% phosphate-buffered formalin, and stained using immunocytochemistry (ICC) with anti-influenza A NP monoclonal antibody (53). A TCID50/ml titer was calculated for each sample using a method described by Reed and Muench (33). To check if all contact pigs seroconverted, blood samples were collected at 14 dpc to perform HI assays (48). The reciprocal titers were divided by 10, log2 transformed, analyzed, and reported as the geometric mean.
Statistical analysis.
Lung lesion scores, log10-transformed virus titers, and log2-transformed HI reciprocal titers were analyzed using t test, with a P value of ≤0.05 considered significant (GraphPad Prism software version 8.00; San Diego, CA).
Data availability.
The sequences from the initial detection of H3.2010.2 are available in GenBank under accession numbers MG720216 and MG720224 (54). GenBank accession numbers for the HA sequence data generated from the study are listed in Table S1. Data used in the study can be accessed through the following link: https://microreact.org/project/4AiXTceagFHQjH7uoZrhUH.
ACKNOWLEDGMENTS
We acknowledge the staff at Iowa State University Veterinary Diagnostic Laboratory for their assistance with diagnostic testing. We also thank Michelle Harland and Nicholas Otis for technical assistance with laboratory techniques and Jason Huegel, Justin Miller, Randy Leon, Adam Hartfiel, and Alyssa Dannen for animal care assistance at USDA, Ames, Iowa.
The study was supported by the Department of Defense, Defense Advanced Research Projects Agency (DARPA), Preventing Emerging Pathogenic Threats program (HR00112020034 to A.S., P.C.G., T.K.A., and A.L.V.) and the SCINet project of the USDA Agricultural Research Service (ARS project number 0500-00093-001-00-D). This study was supported in part by USDA-ARS, USDA-APHIS, an NIH-National Institute of Allergy and Infectious Diseases (NIAID) interagency agreement associated with CRIP (Center of Research in Influenza Pathogenesis), and an NIAID-funded Center of Excellence in Influenza Research and Surveillance (CEIRS, HHSN272201400008C) to A.L.V. C.K.S. was supported by an appointment to the USDA-ARS Research Participation Program administered by the Oak Ridge Institute for Science and Education (ORISE) through an interagency agreement between the U.S. Department of Energy 459 (DOE) and USDA under contract number DE-AC05-06OR23100.
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture, DOE, ORISE, or ISU VDL. USDA is an equal opportunity provider and employer. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Phillip C. Gauger, Email: pcgauger@iastate.edu.
Nicole M. Bouvier, Mount Sinai School of Medicine
REFERENCES
- 1.Bailey ES, Choi JY, Fieldhouse JK, Borkenhagen LK, Zemke J, Zhang D, Gray GC. 2018. The continual threat of influenza virus infections at the human-animal interface: what is new from a one health perspective? Evol Med Public Health 2018:192–198. doi: 10.1093/emph/eoy013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rajao DS, Vincent AL, Perez DR. 2018. Adaptation of human influenza viruses to swine. Front Vet Sci 5:347. doi: 10.3389/fvets.2018.00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deng YM, Wong FYK, Spirason N, Kaye M, Beazley R, Grau MLL, Shan S, Stevens V, Subbarao K, Sullivan S, Barr IG, Dhanasekaran V. 2020. Locally acquired human infection with swine-origin influenza A(H3N2) variant virus, Australia, 2018. Emerg Infect Dis 26:143–147. doi: 10.3201/eid2601.191144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scholtissek C. 1990. Pigs as “mixing vessels” for the creation of new pandemic influenza A viruses. Med Princ Pract 2:65–71. doi: 10.1159/000157337. [DOI] [Google Scholar]
- 5.Johnson NP, Mueller J. 2002. Updating the accounts: global mortality of the 1918–1920 “Spanish” influenza pandemic. Bull Hist Med 76:105–115. doi: 10.1353/bhm.2002.0022. [DOI] [PubMed] [Google Scholar]
- 6.Nelson MI, Vincent AL, Kitikoon P, Holmes EC, Gramer MR. 2012. Evolution of novel reassortant A/H3N2 influenza viruses in North American swine and humans, 2009–2011. J Virol 86:8872–8878. doi: 10.1128/JVI.00259-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ducatez MF, Hause B, Stigger-Rosser E, Darnell D, Corzo C, Juleen K, Simonson R, Brockwell-Staats C, Rubrum A, Wang D, Webb A, Crumpton JC, Lowe J, Gramer M, Webby RJ. 2011. Multiple reassortment between pandemic (H1N1) 2009 and endemic influenza viruses in pigs, United States. Emerg Infect Dis 17:1624–1629. doi: 10.3201/eid1709.110338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson TK, Chang J, Arendsee ZW, Venkatesh D, Souza CK, Kimble JB, Lewis NS, Davis CT, Vincent AL. 2021. Swine influenza A viruses and the tangled relationship with humans. Cold Spring Harb Perspect Med 11:a038737. doi: 10.1101/cshperspect.a038737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson MI, Wentworth DE, Culhane MR, Vincent AL, Viboud C, LaPointe MP, Lin X, Holmes EC, Detmer SE. 2014. Introductions and evolution of human-origin seasonal influenza a viruses in multinational swine populations. J Virol 88:10110–10119. doi: 10.1128/JVI.01080-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nelson MI, Vincent AL. 2015. Reverse zoonosis of influenza to swine: new perspectives on the human-animal interface. Trends Microbiol 23:142–153. doi: 10.1016/j.tim.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zost SJ, Parkhouse K, Gumina ME, Kim K, Diaz Perez S, Wilson PC, Treanor JJ, Sant AJ, Cobey S, Hensley SE. 2017. Contemporary H3N2 influenza viruses have a glycosylation site that alters binding of antibodies elicited by egg-adapted vaccine strains. Proc Natl Acad Sci USA 114:12578–12583. doi: 10.1073/pnas.1712377114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao S, Anderson TK, Walia RR, Dorman KS, Janas-Martindale A, Vincent AL. 2017. The genomic evolution of H1 influenza A viruses from swine detected in the United States between 2009 and 2016. J Gen Virol 98:2001–2010. doi: 10.1099/jgv.0.000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rajão DS, Gauger PC, Anderson TK, Lewis NS, Abente EJ, Killian ML, Perez DR, Sutton TC, Zhang J, Vincent AL. 2015. Novel reassortant human-like H3N2 and H3N1 influenza A viruses detected in pigs are virulent and antigenically distinct from swine viruses endemic to the United States. J Virol 89:11213–11222. doi: 10.1128/JVI.01675-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang J, Anderson TK, Zeller MA, Gauger PC, Vincent AL. 2019. octoFLU: automated classification for the evolutionary origin of influenza a virus gene sequences detected in U.S. swine. Microbiol Resour Announc 8:e00673-19. doi: 10.1128/MRA.00673-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeller MA, Anderson TK, Walia RW, Vincent AL, Gauger PC. 2018. ISU FLUture: a veterinary diagnostic laboratory web-based platform to monitor the temporal genetic patterns of influenza A virus in swine. BMC Bioinformatics 19:397. doi: 10.1186/s12859-018-2408-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walia RR, Anderson TK, Vincent AL. 2019. Regional patterns of genetic diversity in swine influenza A viruses in the United States from 2010 to 2016. Influenza Other Respir Viruses 13:262–273. doi: 10.1111/irv.12559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis NS, Anderson TK, Kitikoon P, Skepner E, Burke DF, Vincent AL. 2014. Substitutions near the hemagglutinin receptor-binding site determine the antigenic evolution of influenza A H3N2 viruses in U.S. swine. J Virol 88:4752–4763. doi: 10.1128/JVI.03805-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nelson MI, Gramer MR, Vincent AL, Holmes EC. 2012. Global transmission of influenza viruses from humans to swine. J Gen Virol 93:2195–2203. doi: 10.1099/vir.0.044974-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Webby RJ, Rossow K, Erickson G, Sims Y, Webster R. 2004. Multiple lineages of antigenically and genetically diverse influenza A virus co-circulate in the United States swine population. Virus Res 103:67–73. doi: 10.1016/j.virusres.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 20.Zhou NN, Senne DA, Landgraf JS, Swenson SL, Erickson G, Rossow K, Liu L, Yoon K-J, Krauss S, Webster RG. 1999. Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J Virol 73:8851–8856. doi: 10.1128/JVI.73.10.8851-8856.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Webby RJ, Swenson SL, Krauss SL, Gerrish PJ, Goyal SM, Webster RG. 2000. Evolution of swine H3N2 influenza viruses in the United States. J Virol 74:8243–8251. doi: 10.1128/jvi.74.18.8243-8251.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bolton MJ, Abente EJ, Venkatesh D, Stratton JA, Zeller M, Anderson TK, Lewis NS, Vincent AL. 2019. Antigenic evolution of H3N2 influenza A viruses in swine in the United States from 2012 to 2016. Influenza Other Respir Viruses 13:83–90. doi: 10.1111/irv.12610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wagner R, Matrosovich M, Klenk HD. 2002. Functional balance between haemagglutinin and neuraminidase in influenza virus infections. Rev Med Virol 12:159–166. doi: 10.1002/rmv.352. [DOI] [PubMed] [Google Scholar]
- 24.Steel J, Lowen AC, Mubareka S, Palese P. 2009. Transmission of influenza virus in a mammalian host is increased by PB2 amino acids 627K or 627E/701N. PLoS Pathog 5:e1000252. doi: 10.1371/journal.ppat.1000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chou YY, Albrecht RA, Pica N, Lowen AC, Richt JA, García-Sastre A, Palese P, Hai R. 2011. The M segment of the 2009 new pandemic H1N1 influenza virus is critical for its high transmission efficiency in the guinea pig model. J Virol 85:11235–11241. doi: 10.1128/JVI.05794-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rajão DS, Walia RR, Campbell B, Gauger PC, Janas-Martindale A, Killian ML, Vincent AL. 2017. Reassortment between swine H3N2 and 2009 pandemic H1N1 in the United States resulted in influenza A viruses with diverse genetic constellations with variable virulence in pigs. J Virol 91:e01763-16. doi: 10.1128/JVI.01763-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelson MI, Souza CK, Trovão NS, Diaz A, Mena I, Rovira A, Vincent AL, Torremorell M, Marthaler D, Culhane MR. 2019. Human-origin influenza A(H3N2) reassortant viruses in swine, southeast Mexico. Emerg Infect Dis 25:691–700. doi: 10.3201/eid2504.180779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abente EJ, Santos J, Lewis NS, Gauger PC, Stratton J, Skepner E, Anderson TK, Rajao DS, Perez DR, Vincent AL. 2016. The molecular determinants of antibody recognition and antigenic drift in the H3 hemagglutinin of swine influenza A virus. J Virol 90:8266–8280. doi: 10.1128/JVI.01002-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Das SR, Hensley SE, Ince WL, Brooke CB, Subba A, Delboy MG, Russ G, Gibbs JS, Bennink JR, Yewdell JW. 2013. Defining influenza A virus hemagglutinin antigenic drift by sequential monoclonal antibody selection. Cell Host Microbe 13:314–323. doi: 10.1016/j.chom.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altman MO, Angel M, Košík I, Trovão NS, Zost SJ, Gibbs JS, Casalino L, Amaro RE, Hensley SE, Nelson MI, Yewdell JW. 2019. Human influenza A virus hemagglutinin glycan evolution follows a temporal pattern to a glycan limit. mBio 10:e00204-19. doi: 10.1128/mBio.00204-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bowman AS, Walia RR, Nolting JM, Vincent AL, Killian ML, Zentkovich MM, Lorbach JN, Lauterbach SE, Anderson TK, Davis CT, Zanders N, Jones J, Jang Y, Lynch B, Rodriguez MR, Blanton L, Lindstrom SE, Wentworth DE, Schiltz J, Averill JJ, Forshey T. 2017. Influenza A(H3N2) virus in swine at agricultural fairs and transmission to humans, Michigan and Ohio, USA, 2016. Emerg Infect Dis 23:1551–1555. doi: 10.3201/eid2309.170847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duwell MM, Blythe D, Radebaugh MW, Kough EM, Bachaus B, Crum DA, Perkins KA, Jr, Blanton L, Davis CT, Jang Y, Vincent A, Chang J, Abney DE, Gudmundson L, Brewster MG, Polsky L, Rose DC, Feldman KA. 2018. Influenza A(H3N2) variant virus outbreak at three fairs–Maryland, 2017. MMWR Morb Mortal Wkly Rep 67:1169–1173. doi: 10.15585/mmwr.mm6742a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reed L, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Epidemiol 27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- 34.Chen Q, Wang L, Zheng Y, Zhang J, Guo B, Yoon KJ, Gauger PC, Harmon KM, Main RG, Li G. 2018. Metagenomic analysis of the RNA fraction of the fecal virome indicates high diversity in pigs infected by porcine endemic diarrhea virus in the United States. Virol J 15:95. doi: 10.1186/s12985-018-1001-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J, Zheng Y, Xia XQ, Chen Q, Bade SA, Yoon KJ, Harmon KM, Gauger PC, Main RG, Li G. 2017. High-throughput whole genome sequencing of porcine reproductive and respiratory syndrome virus from cell culture materials and clinical specimens using next-generation sequencing technology. J Vet Diagn Invest 29:41–50. doi: 10.1177/1040638716673404. [DOI] [PubMed] [Google Scholar]
- 36.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. 2009. ABySS: a parallel assembler for short read sequence data. Genome Res 19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, Aevermann BD, Anderson TK, Burke DF, Dauphin G, Gu Z, He S, Kumar S, Larsen CN, Lee AJ, Li X, Macken C, Mahaffey C, Pickett BE, Reardon B, Smith T, Stewart L, Suloway C, Sun G, Tong L, Vincent AL, Walters B, Zaremba S, Zhao H, Zhou L, Zmasek C, Klem EB, Scheuermann RH. 2017. Influenza Research Database: an integrated bioinformatics resource for influenza virus research. Nucleic Acids Res 45:D466–D474. doi: 10.1093/nar/gkw857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Price MN, Dehal PS, Arkin AP. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A. 2018. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol 4:vey016. doi: 10.1093/ve/vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koel BF, Burke DF, Bestebroer TM, van der Vliet S, Zondag GCM, Vervaet G, Skepner E, Lewis NS, Spronken MIJ, Russell CA, Eropkin MY, Hurt AC, Barr IG, de Jong JC, Rimmelzwaan GF, Osterhaus ADME, Fouchier RAM, Smith DJ. 2013. Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science 342:976–979. doi: 10.1126/science.1244730. [DOI] [PubMed] [Google Scholar]
- 47.Rajao DS, Anderson TK, Kitikoon P, Stratton J, Lewis NS, Vincent AL. 2018. Antigenic and genetic evolution of contemporary swine H1 influenza viruses in the United States. Virology 518:45–54. doi: 10.1016/j.virol.2018.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kitikoon P, Gauger PC, Vincent AL. 2014. Hemagglutinin inhibition assay with swine sera. Methods Mol Biol 1161:295–301. doi: 10.1007/978-1-4939-0758-8_24. [DOI] [PubMed] [Google Scholar]
- 49.R Core Development Team. 2019. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 50.Wickham H. 2016. ggplot2: elegant graphics for data analysis. Springer-Verlag; New York, NY. [Google Scholar]
- 51.Gauger PC, Vincent AL, Loving CL, Henningson JN, Lager KM, Janke BH, Kehrli ME, Jr, Roth JA. 2012. Kinetics of lung lesion development and pro-inflammatory cytokine response in pigs with vaccine-associated enhanced respiratory disease induced by challenge with pandemic (2009) A/H1N1 influenza virus. Vet Pathol 49:900–912. doi: 10.1177/0300985812439724. [DOI] [PubMed] [Google Scholar]
- 52.Vincent AL, Lager KM, Janke BH, Gramer MR, Richt JA. 2008. Failure of protection and enhanced pneumonia with a US H1N2 swine influenza virus in pigs vaccinated with an inactivated classical swine H1N1 vaccine. Vet Microbiol 126:310–323. doi: 10.1016/j.vetmic.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 53.Kitikoon P, Nilubol D, Erickson BJ, Janke BH, Hoover TC, Sornsen SA, Thacker EL. 2006. The immune response and maternal antibody interference to a heterologous H1N1 swine influenza virus infection following vaccination. Vet Immunol Immunopathol 112:117–128. doi: 10.1016/j.vetimm.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 54.Zeller MA, Li G, Harmon KM, Zhang J, Vincent AL, Anderson TK, Gauger PC. 2018. Complete genome sequences of two novel human-like H3N2 influenza A viruses, A/swine/Oklahoma/65980/2017 (H3N2) and A/Swine/Oklahoma/65260/2017 (H3N2), detected in swine in the United States. Microbiol Resour Announc 7:e01203-18. doi: 10.1128/MRA.01203-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Maximum likelihood (ML) phylogenetic trees. (A) ML phylogenetic tree of the swine H3-HA clades currently circulating in swine. The novel H3.2010.2 HA clade is highlighted in blue. (B) ML phylogenetic tree of the swine N2-NA clades currently circulating in swine. H3.2010.2 NA clade is highlighted in blue. The branches are colored based on phylogenetic clade designation. Download FIG S1, TIF file, 0.2 MB (162.5KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Swine influenza A virus cases detected in the ISU VDL from the recent sustained human to swine spillover from 2017 to 2020 in the United States. Download Table S1, PDF file, 0.04 MB (41.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Representative H3.2010.1 and H3.2010.2 influenza A virus from swine used in the antigenic distance analysis. Download Table S2, PDF file, 0.01 MB (13.1KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
HA1 percentage of amino acid identity of H3.2010.1 and H3.2010.2 strains used in the antigenic distance analysis. Download Table S4, PDF file, 0.01 MB (13.9KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Cross hemagglutination inhibition (HI) titers representing influenza A viruses from the antigenic map and antigenic distance analysis. Download Table S5, PDF file, 0.02 MB (22.1KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Hemagglutinin (HA) amino acid sequence alignment of the H3.2010.1 and H3.2010.2 swine lineages used for antigenic analysis. Identity to the consensus sequences is represented by a dot. Antigenic motif (H3 numbering: 145, 155, 156, 158, 159, 189) is highlighted with red boxes. Download Table S3, PDF file, 0.2 MB (186.9KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data Availability Statement
The sequences from the initial detection of H3.2010.2 are available in GenBank under accession numbers MG720216 and MG720224 (54). GenBank accession numbers for the HA sequence data generated from the study are listed in Table S1. Data used in the study can be accessed through the following link: https://microreact.org/project/4AiXTceagFHQjH7uoZrhUH.