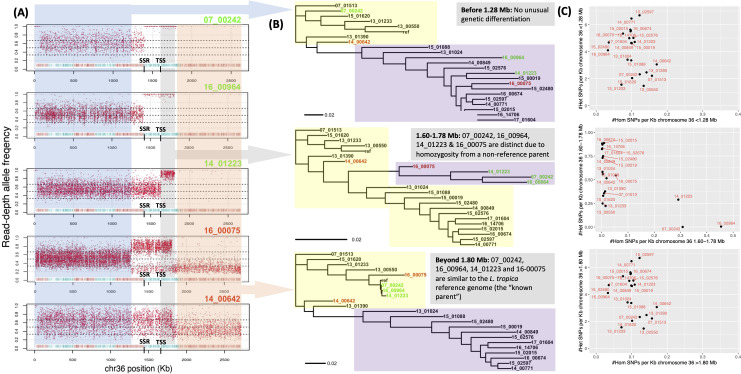
Fig 3.
RDAF distributions (A) and phylogenies (B) constructed from SNPs at the 5’-end of chromosome 36 (<1280 Kb, blue RDAF, top phylogeny), middle (1600–1780 Kb, grey RDAF, middle phylogeny) and 3’ end (>1800 Kb, orange RDAF, bottom phylogeny) showing the genetic relatedness of the 22 isolates with the L. tropica reference genome (“ref”). (A) Patterns of heterozygosity (blue), a LROH (grey) and homozygous similarity (orange) are seen in 14_01223, 07_00242 and 16_00964 (green), and high heterozygosity in 16_00075 and 14_642 (red). All isolates were disomic for chromosome 36, except 14_00642 that was trisomic. The putative TSS inferred from L. major is at 1.63 Mb (black), corresponding to a recombination breakpoint in 14_01223 (middle). 07_00242 (top) and 16_00964 (second) had breakpoint at an inferred strand-switch region (SSR) at 1.42 Mb. (B) The inferred genetically distinct groups are represented by the L. tropica reference (yellow area) and non-reference groups (purple area). (C) The homozygous (x-axis) and heterozygous (y-axis) SNPs/Kb for all 22 samples for the 0–1.28 Mb region (top), 1.60–1.78 region (middle) and >1.8 Mb region (bottom).