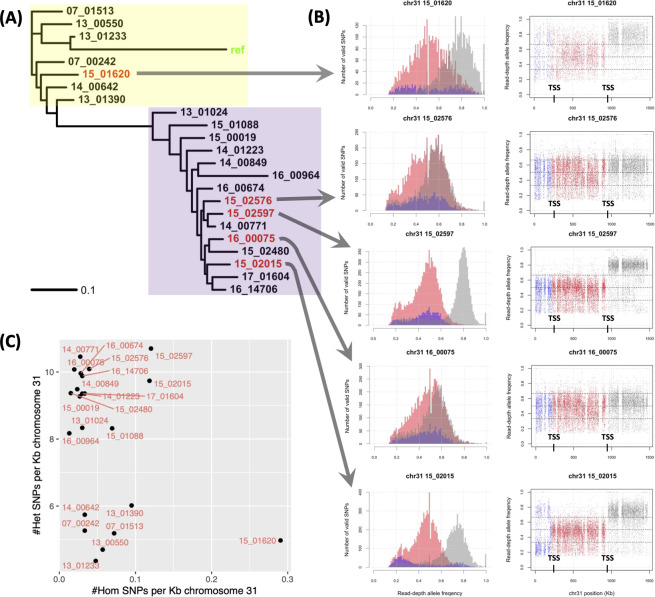
Fig 4. Heterozygosity was high and haplotype-specific on chromosome 31.
(A) A rooted phylogeny constructed from SNPs at chromosome 31 showing a divergent L. tropica reference (“ref”, green) due to a lack of heterozygous SNPs in this haploid genome. The inferred genetically distinct groups are represented by a group more related to the L. tropica reference (yellow area) and the non-reference group (purple area). (B) The read-depth allele frequency (RDAF) SNP densities (left) and distribution (right) across chromosome 31 (left) showed distinct patterns at the 5’ end (<220 Kb, blue), middle (220–950 Kb, red) and 3’ end (>950 Kb). 15_01620 (top) and 15_02015 (bottom) had recombination breakpoints at 220 and 950 Kb, coinciding with two putative TSSs inferred from experiments in L. major were at 220 and 950 Kb (black). 15_02597 (middle) also had a breakpoint at 950 Kb. 15_02576 (second) and 16_00075 (fourth) had no clear breakpoints. (C) The homozygous (x-axis) and heterozygous (y-axis) SNPs/Kb.