Fig. 5. REV-ERBs control cardiac NAD+ biosynthesis via E4BP4-mediated repression of Nampt.
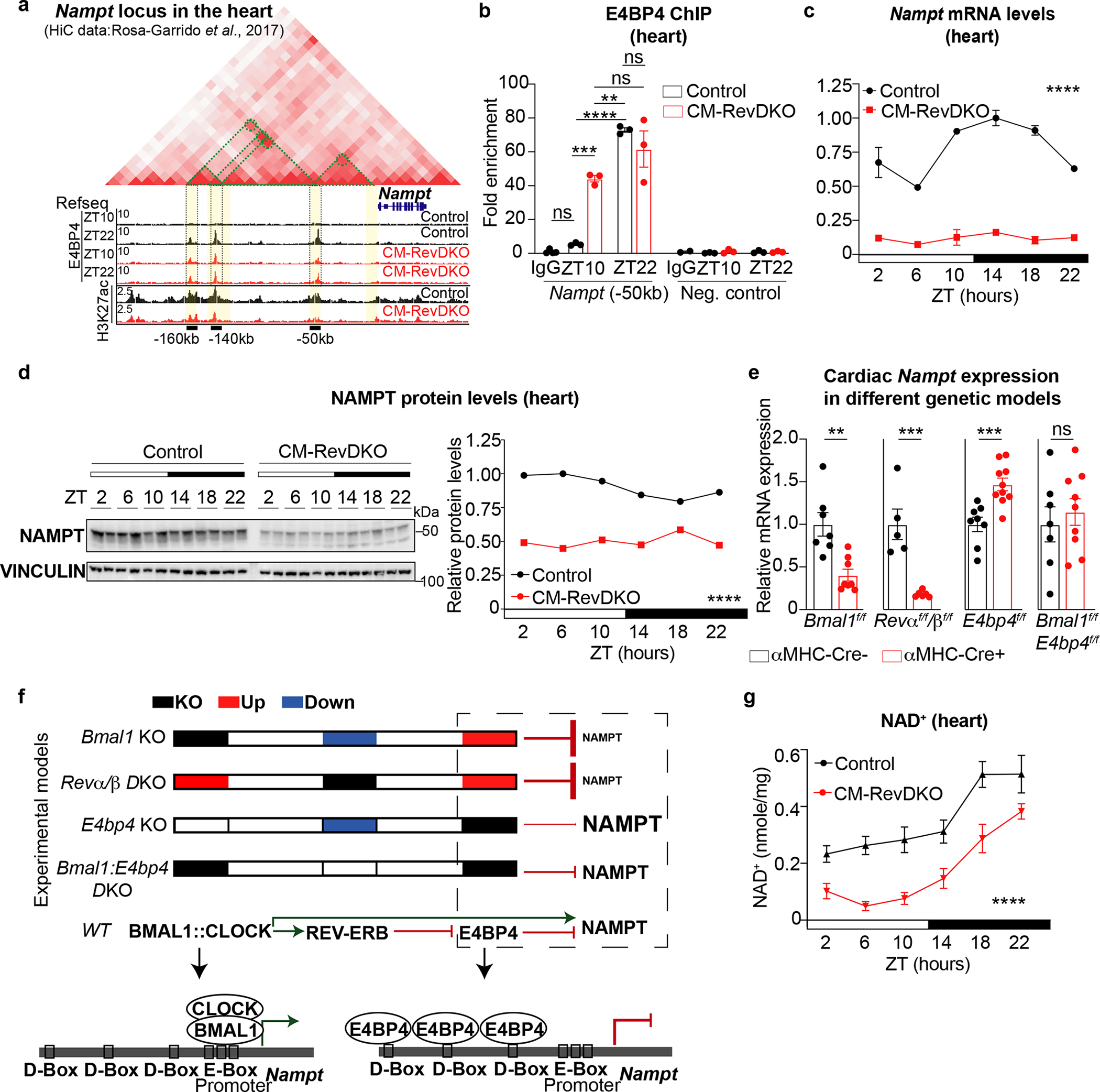
a, Hi-C from the heart (obtained from39), H3K27ac Cut&Run data from isolated adult control and CM-RevDKO hearts (at ZT10) and E4BP4 ChIP-Seq data (at ZT10 and ZT22) from control and CM-RevDKO hearts. Green dotted lines delineate 3D contacts. Identified cis-regulatory elementals (CREs) are highlighted in yellow, 3 of them were found to be bound by E4BP4, marked by black bars and their respective distance to the TSS of Nampt. Data were visualized via Juicebox64. b, E4BP4 ChIP-qPCR at the CRE, identified in (a), 50kb upstream of the Nampt TSS (at ZT10 and ZT22) from control and CM-RevDKO hearts. A random locus in the genome was chosen as negative control. (n = 3/condition/genotype). c, Relative Nampt mRNA expression in control vs CM-RevDKO hearts from 2-month-old male mice (n = 3 hearts/genotype/timepoint except for n = 2 for ZT6/ZT10 and ZT22 in control, ZT2 and ZT18 in CM-RevDKO, and n = 4 for ZT22 in CM-RevDKO). d, Immunoblot and protein quantification for NAMPT from control vs CM-RevDKO hearts from 2-month-old male mice (n=2/timepoint/genotype). e, Relative Nampt mRNA expression in hearts from multiple genetic cardiomyocyte-specific (CM) KO models: CBK (n = 7 for control and n = 8 for KO), CM-RevDKO (n = 5 for control and n = 6 for KO), E4bp4 (n = 8 for control and n = 10 for KO) and CBK/E4bp4 (double) KO (n = 6 for control and n = 9 for DKO) and control hearts from 2-month-old male mice for CM-RevDKO and control at ZT10 and from 3-month-old male mice for the rest at ZT12. f, cartoon of summarized experimental data and proposed mechanism of transcriptional regulation of Nampt in different genetic KO models. g, NAD+ levels from control vs CM-RevDKO hearts from 2-month-old mice (n = 6 hearts/timepoint except for n = 5 for ZT6 in control, n = 5 except for n = 4 for ZT6, n = 6 for ZT18 and n = 7 for ZT22 in CM-RevDKO). n represents biologically independent replicates. All data are presented as mean ± SEM, except for d. ns: non significant, **P < 0.01,***P < 0.001,****P < 0.0001 by two-way ANOVA (in c,d,g), one-way ANOVA followed by a Tukey’s multiple comparisons test (in b), and by 2-sided Student’s t test in e (exact P values are provided in the Source Data).
