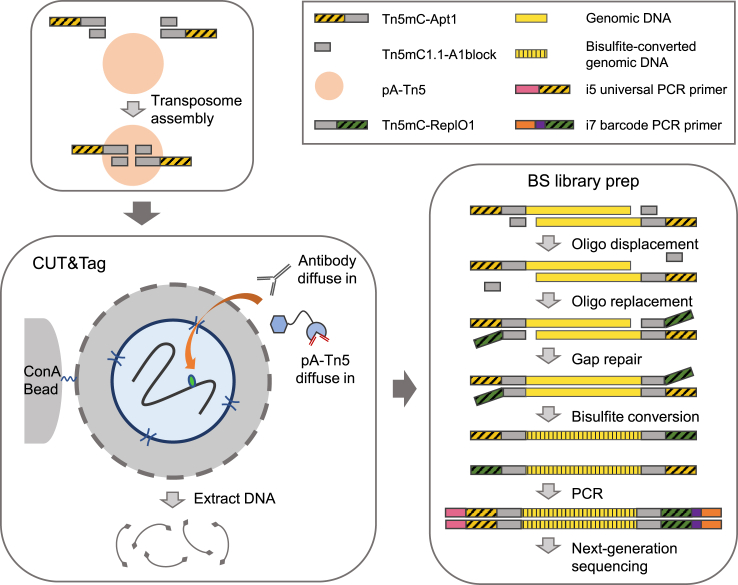
Figure 1.
A schematic overview of CUT&Tag-BS workflow
CUT&Tag is adapted to couple with tagmentation-based bisulfite sequencing by using pA-Tn5 assembled with a methylated adapter. The top adapter, Tn5mC-Apt1, and the replacement oligonucleotide, Tn5mC-ReplO1, must be methylated at all cytosines to maintain their identity during bisulfite treatment. CUT&Tag is performed according to the published protocol (Kaya-Okur et al., 2019) with pA-Tn5 loaded with a methylated adapter. Briefly, cells are harvested and bound to concanavalin A-coated magnetic beads. The cell membrane is permeabilized with digitonin (indicated by holes in the membrane) to allow the antibodies and pA-Tn5 to diffuse into the cells to find their targets and then tagment genomic binding sites of the target protein. Tagmented DNA from CUT&Tag is subsequently subjected to tagmentation-based bisulfite sequencing library preparation. The oligonucleotide replacement and gap-repair step covalently attach the methylated adapter Tn5mC-ReplO1 to each DNA strand, followed by bisulfite conversion and PCR amplification to generate the library for sequencing. This figure is adapted from previously published figures in Skene et al. (2018) and Wang et al. (2013) by permission from Springer Nature.