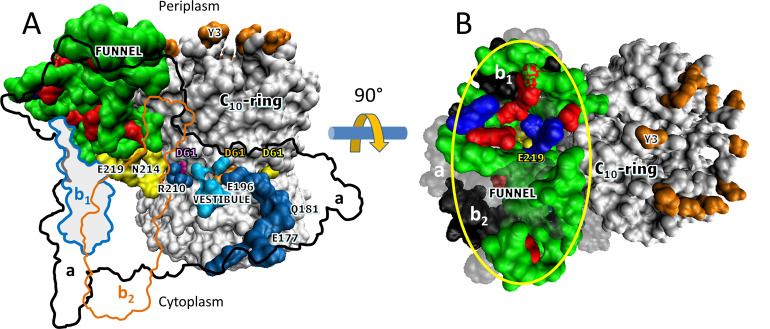
Figure 5. Aqueous funnel of charged and polar groups can serve as an antenna to supply protons to the input channel.
(A) Transmembrane view of Escherichia coli FO (pdb-ID 6OQR) showing the path of charged and polar residues across the membrane. Outlines indicate space occupied by hydrophobic residues in subunit-a (black line), subunit-b1 (blue line), and subunit-b2 (orange line). The inner surface of the funnel, which is lined with polar residues and loop regions (green), acidic groups (red), and histidines (blue) from subunit-a and the subunit-b1 N-terminus, is exposed to the periplasm at its wide end that narrows to aE219 (yellow) at the bottom. The input channel (yellow) extends from aE219 to aN214 and aQ252, which are proximal to aR210. Between aR210 and the output channel (dark blue) the leading (pink) and lagging (orange) cD61 groups rotate through a vestibule lined above and below the cD61 rotation plane by polar sidechains (light blue) that decrease the dielectric constant of the vestibule from that of the lipid bilayer. A protonated cD61 exposed to the lipid bilayer (yellow) is also visible. (B) Periplasmic surface of FO showing the interior surface of the funnel (orange oval) lined with charged and polar groups from subunit-a and subunit-b1 as in A that narrows to aE219 (yellow) at the bottom where the input channel begins. Hydrophobic residues are shown of subunit-a (gray), subunits-b1 and -b2 (black), and the c-ring (white). The cY3 sidechains (orange) are shown to indicate the orientation of the periplasmic surface of the c-ring.