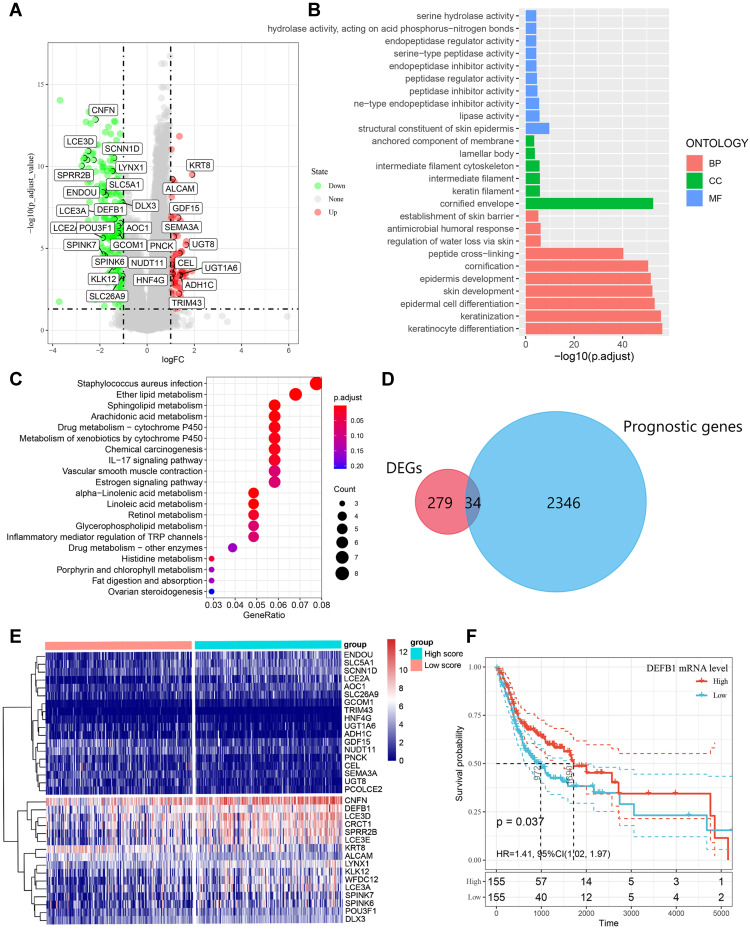
Figure 4.
Identification of differentially expressed genes (DEGs) that related with CD4+ central memory T cell and functional enrichment analysis. (A) The volcano plot of DEGs between oral squamous cell carcinoma (OSCC) groups with high and low CD4+ central memory T cell abundance. (B)Gene Ontology (GO) enrichment analysis of DEGs related with CD4+ central memory T cell. MF, molecular function; CC, cell component; BP, biological process. (C) Enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways of DEGs related with CD4+ central memory T cell. (D) Venn diagram of the overlapped genes between DEGs related with CD4+ central memory T cell and prognostic genes in OSCC patients. (E) Cluster analysis for prognostic DEGs related with CD4+ central memory T cell OSCC patients from the TCGA cohort. (F) The survival rate of OSCC patients in the TCGA cohort with high and low DEFB1 expression.