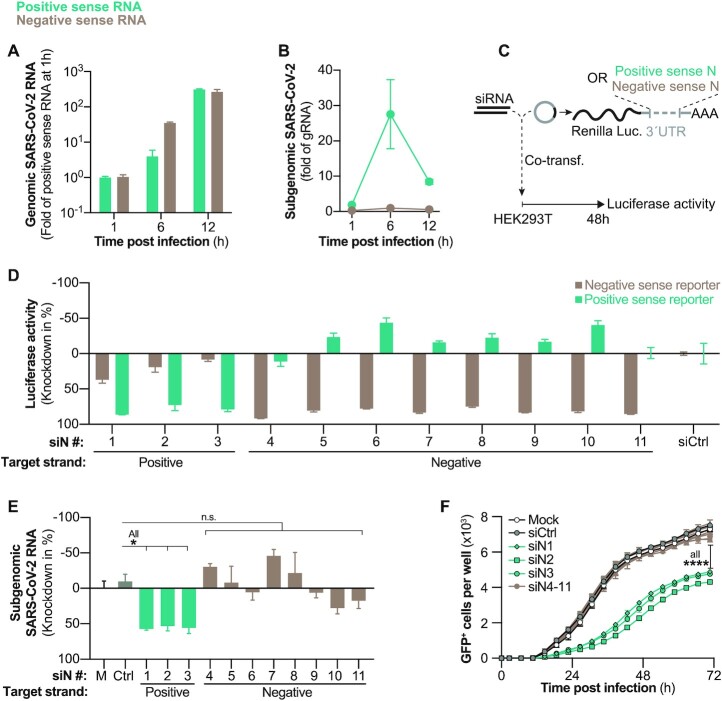
Figure 2.
Evaluation of SARS-CoV-2 negative sense RNA as siRNA target. (A, B) Kinetics of negative and positive sense SARS-CoV-2 RNAs following wildtype SARS-CoV-2 infection (MOI 0.1) of VeroE6 cells. Negative and positive sense RNAs were individually transcribed to cDNA by using either poly A or poly T primers and (A) gRNA and (B) sgRNAs quantified by RT-qPCR. (C) Experimental setup to determine siRNA strand specific activities. Luciferase reporters with incorporated positive or negative sense N sequences in the 3′UTR of Renilla luciferase were co-transfected with siRNAs into HEK293T cells and (D) luciferase activity measured after 48 h (E) siRNAs were transfected into VeroE6 cells 6 h before infection with wildtype SARS-CoV-2 (MOI 0.1) and 24 h p.i. sgRNAs quantified from cell lysate using RT-qPCR. (F) Same setup as in (E) but VeroE6 cells were infected with rSARS-CoV-2-GFP (MOI 1.0) and GFP+ cells quantified every 4 h. All experiments were performed with three biological replicates. Graphs in (A, B, D, E) show mean and error bars SEM. Statistical differences were calculated using (E) Regular or (F) repeated measures one-way Anova with Dunnett′s multiple comparison correction. Co-transf., co-transfection; M = mock-transfected; n.s., non-significant, *P < 0.05; ****P < 0.0001