Figure 1.
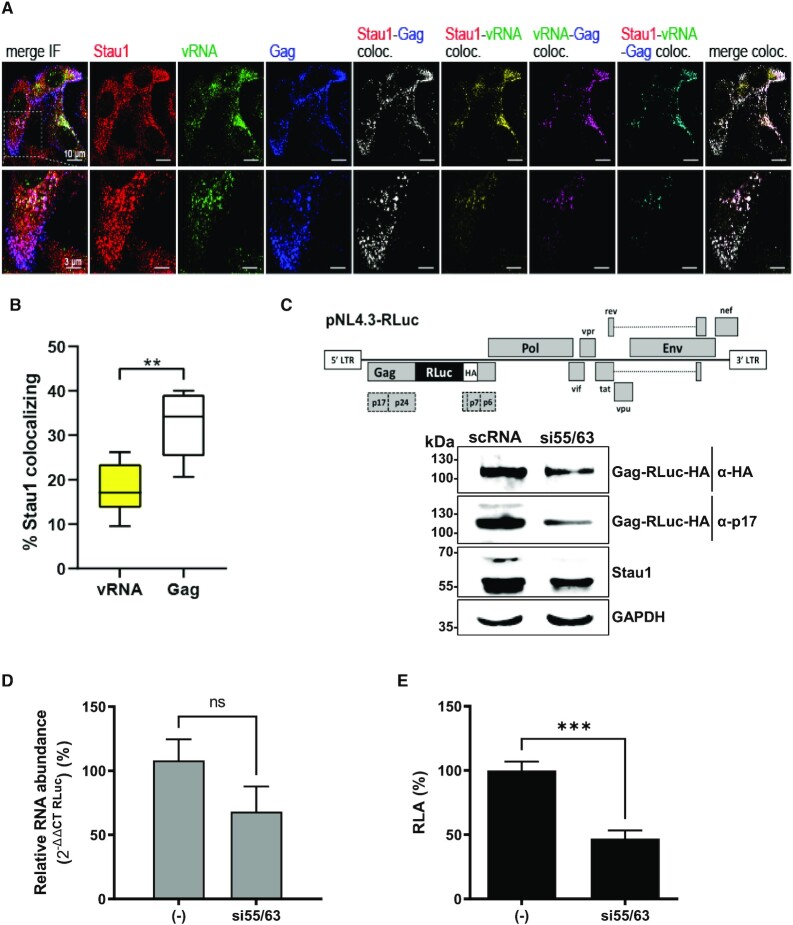
Staufen1 binds the vRNA and regulated HIV-1 Gag expression. (A) HeLa cells were transfected with pNL-4.3 proviral DNA and at 48 h post-transfection, combined IF/FISH was performed as described in Materials and Methods for endogenous Staufen1 (red), HIV-1 vRNA (green), and HIV-1 Gag (blue). A merged panel showing colocalization Staufen1, HIV-1 vRNA and HIV-1 Gag signals is shown on the far left (last panel), while resulting colocalization channels between Staufen1-Gag (white), Staufen1-vRNA (yellow), vRNA-Gag (magenta), and Staufen1-vRNA-Gag (cyan), as determined using Imaris software, are shown, as indicated. (B) Imaris was used to determine percent colocalization between Staufen1 and vRNA (17.70% ± 5.79 S.D.; yellow bar) and Staufen1 and Gag (32.07% ±7.38 S.D., white bar). Statistical analyses were performed by an unpaired two-tailed t-test (P = 0.0016). (C) Schematic representation of the complete HIV-1 molecular clone pNL-4.3-RLuc (upper panel). HEK 293T cells were cotransfected with the pNL-4.3-RLuc (200 ng) plasmid and 50 nM of a duplex siRNA targeting the Staufen1 mRNA (si55/63), or with a scrambled RNA ((−); 50 nM) as a control. The expression of the HIV-1 Gag-RLuc-HA fusion protein and endogenous Staufen1 was determined 48 h post-transfection by western blot, using the GAPDH protein as a loading control. (D) Cytoplasmic RNA was extracted from cells expressing pNL-4.3-RLuc HIV-1 and treated with the scRNA or the si55/63 (50 nM), and relative RNA levels were determined by real-time RT-qPCR. The RNA abundance was expressed relative to the value obtained for the cells treated with the scRNA set to 100%. (E) Renilla luciferase activity was measured 48 h post-transfection and is expressed as relative luciferase activity (RLA) relative to the activity obtained when the pNL-4.3-RLuc plasmid was cotransfected with the scRNA (−). In (D) and (E) values represent the mean (±SEM) for seven independent experiments, each conducted in duplicate. Statistical analyses were performed by an unpaired two-tailed t-test (* P < 0.01; *** P < 0.0005).