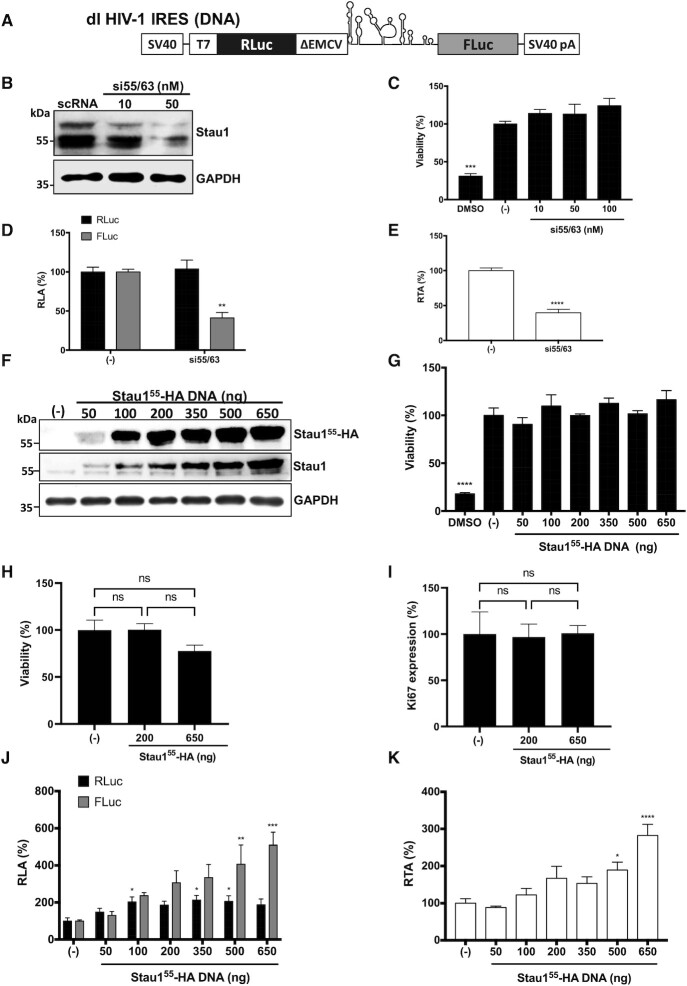
Figure 2.
Staufen1 is an ITAF for the HIV-1 IRES. (A) Schematic representation of dl HIV-1 IRES that harbors the 5′UTR of the HIV-1 mRNA (pNL-4.3 HIV-1 clone, GenBank accession number AF 324493) in the intercistronic spacer flanked by RLuc and FLuc ORFs (16). (B–E) The dl HIV-1 IRES plasmid (300 ng) was cotransfected with the si55/63 targeting Staufen mRNA (10–50 nM) or with the scRNA ((−); 50 nM) in HEK 293T cells. (B) The expression level of endogenous Stau155 and Stau163 was determined by western blot using an anti-Staufen1 antibody and GAPDH as a loading control. (C) Cell viability was determined by measuring the cellular metabolic activity using the MTS assay using dimethylsulfoxide (DMSO, 10%) as a control for cell death. Data are expressed relative to the viability of the cells transfected with scRNA(−) set to 100%. Values shown are the mean (±SEM) for three independent experiments, each performed in duplicate. Statistical analysis was performed by an ordinary one-way ANOVA test (***P < 0.001). (D, E) RLuc and Fluc activities were measured 48 h post-transfection, and data are presented as RLA (D) and relative translational activity (RTA) (E), with the values obtained in the presence of the scRNA (−) set to 100%. RTA corresponds to the [FLuc/RLuc] ratio that is used as an index of IRES activity. Values represent the mean (±SEM) for three independent experiments, each conducted in duplicate. Statistical analysis was performed by an ordinary two-way ANOVA test (** P < 0.005, **** P < 0.0001). (F–K) HEK 293T cells were cotransfected with the dl HIV-1 IRES (300 ng) and different quantities (50–650 ng) of a plasmid encoding for a Stau155-HA3 protein. The expression of Stau155-HA3 was confirmed by western blot using an anti-HA or anti-Staufen1 antibody and GAPDH as a loading control. (G, H) The viability of HEK 293T cells was determined either by measuring the cellular metabolic activity using the MTS assay and dimethylsulfoxide (DMSO, 10%) as a control for cell death (G) or by using flow cytometry (FC) in cells stained with Fixed Viability Stain 510 and determining the (5) of living cells (H). In (G, H), data are expressed relative to the viability of the cells transfected with the negative control set to 100%. (I) Cell proliferation was analyzed by detecting Ki67 protein in HEK 293T cells transfected or not with the Stau155-HA3 expressing plasmid (200 or 650 ng). For (G–I) values are the mean (±SEM) for three independent experiments, each performed in duplicate. Statistical analysis was performed by an ordinary one-way ANOVA test (**P < 0.005, ****P < 0.0001; ns, not significant). (J, K). RLuc and FLuc activities were measured 24 h post-transfection, and data are presented RLA (J) or RTA (K). The RLA and RTA values obtained in the absence (−) of the Stau155-HA3 plasmid were set to 100%. Values shown are the mean (±SEM) of four independent experiments, each performed in duplicate. Statistical analysis was performed by an ordinary one-way ANOVA test (* P < 0.05, ** P < 0.005, ***P < 0.001, ****P < 0.0001).