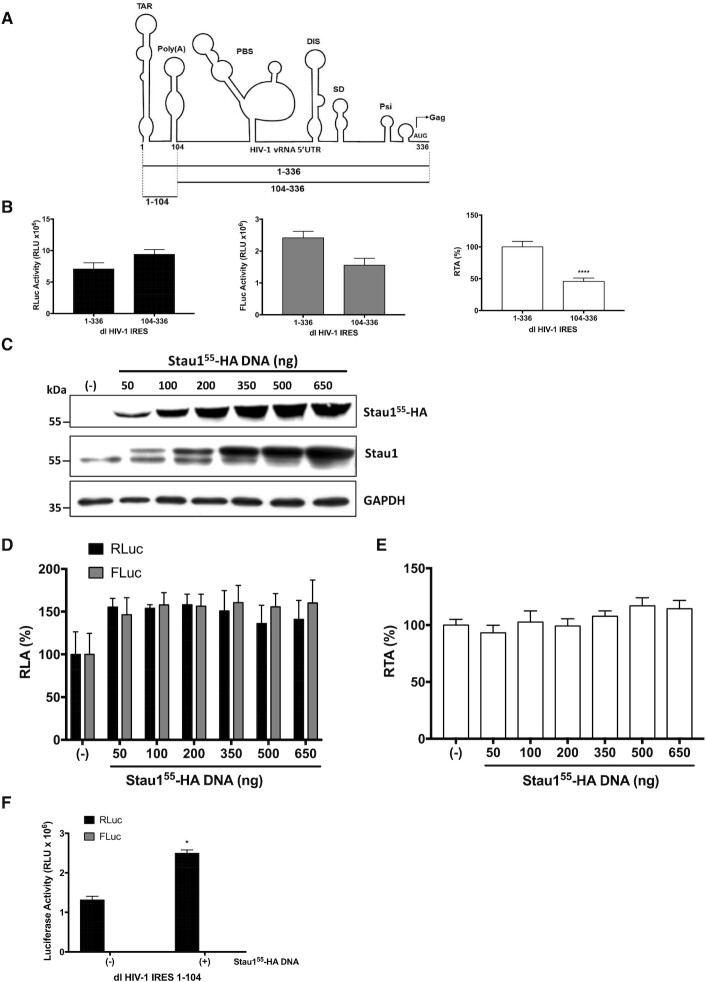
Figure 4.
Overexpression of Staufen1 does not stimulate the activity of a 5′deletion mutant of the HIV-1 IRES lacking the TAR and Poly(A) stem-loops. (A) Schematic representation of the pNL-4.3 vRNA 5′UTR used to generate dl HIV-1 plasmids (intercistronic region), dl HIV-1 IRES (nts 1–336), dl HIV-1 IRES 104–336 (nts 104–336), and dl HIV-1 IRES 1–104 (nts 1–104). The different RNA structural elements (TAR, poly(A) loop, PBS, DIS, SD and Psi) present within the HIV-1 vRNA 5′UTR are depicted (21). The arrow at position 336 indicated the Gag ORF initiation codon. (B) HEK 293T cells were transfected with the dl HIV-1 IRES or dl HIV-1 IRES 104–336 (300 ng) plasmids. RLuc (left panel) and FLuc (middle panel) activities were measured 24 h post-transfection, and data are presented as relative light units (RLU) or as RTA (right panel). (C−E) HEK 293T cells were cotransfected with the dl HIV-1 IRES 104–336 (300 ng) and different quantities (50–650 ng) of a plasmid encoding for a Stau155-HA3. (C) The presence of the overexpressed Stau155-HA3 was confirmed by western blot using an anti-HA or anti-Satufen1 antibody and GAPDH as a loading control. (D, E) RLuc and FLuc activities were measured 24 h post-transfection, and data are presented as RLA (D) or RTA (E). The RLA and RTA values obtained in the absence (−) of the Stau155-HA plasmid were set to 100%. Values shown are the mean (+/-SEM) of four independent experiments, each performed in duplicate. Statistical analysis was performed by an ordinary one-way ANOVA test. (F) HEK 293T cells were cotransfected with the dl HIV-1 IRES 1–104 (300 ng) and 650 ng of a plasmid encoding for a Stau155-HA3 protein. RLuc and FLuc activities were measured 24 h post-transfection, and data are presented as RLU. Values shown are the mean (−SEM) of three independent experiments, each performed in duplicate. Statistical analysis was performed by an ordinary one-way ANOVA test (* P < 0.05).