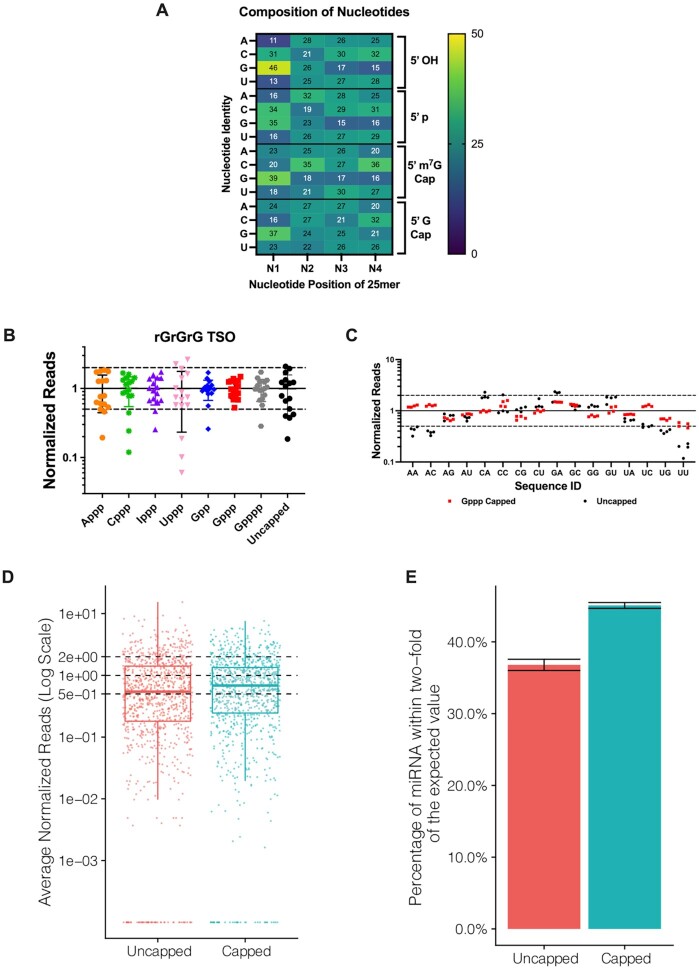
Figure 3.
Chemical capping reduces template switching biases. (A) Heat map of the contribution of the first four nucleotides (N1–N4) and the RNA 5′ end capping to template switching bias. Color gradient indicates normalized representation of nucleotide N in the sequencing reads: yellow (overrepresented), dark blue (underrepresented) and teal (neutral). The presence of an unmethylated guanosine cap minimizes overall nucleotide representation bias in the library. Heat map data represent means of n = 3–4 independent experiments, ±SD < 1. (B) Normalized read distribution of libraries prepared from a pool of 16 discrete RNA templates varying the first two nucleotides N1 and N2. The RNA pool was subjected to chemical capping as indicated. Template switching was performed with the rGrGrG-3′ TSO. Reads were normalized to 1 (solid line) and the interval of 2-fold above and below this boundary was denoted by dashed lines. (C) 5′-Gppp capped or uncapped RNA libraries shown in (B) arranged by the identity of the first two 5′ end nucleotides. Data shown in (B) and (C) represent mean ± SD of n = 4 experiments. Assays were performed with 25mer RNAs, either comprising synthetic oligonucleotides with randomized positions (A) or a pool of discrete sequences (B and C). (D) Normalized read counts for 0.5 pg input uncapped and capped libraries prepared from an equimolar pool of 962 synthetic miRNAs (miRXplore reference). Each miRNA is expected to have a normalized read value of 1 (central dashed line). The upper and lower dashed lines correspond to the interval of 2-fold above and below the expected value. (E) Percentage of miRNAs from (D) falling within 2-fold of their expected values shown as the average of three technical replicates per method. Error bars represent the standard deviation.