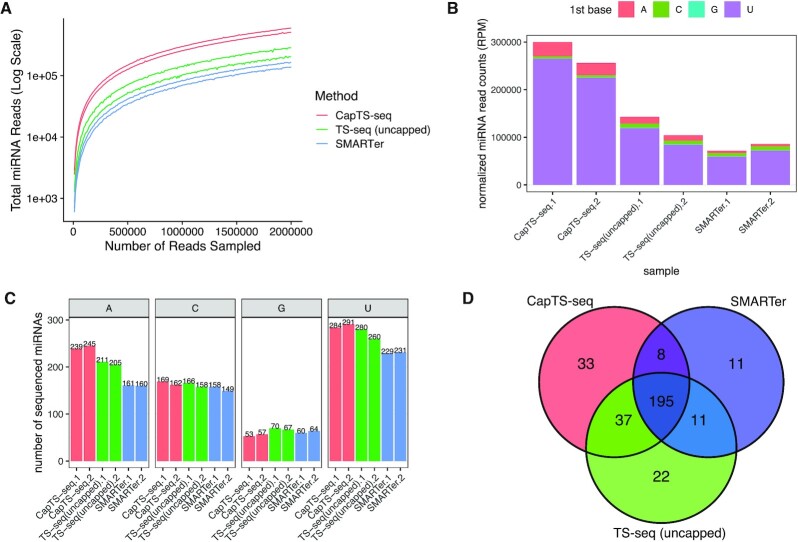
Figure 5.
CapTS-seq improves miRNA detection in total brain RNA. (A) Number of miRNAs detected with a mirDeep2 score >0 at various subsampled read depths. Datasets were randomly sampled with the number of reads increasing by 5000 in each sample. Values represent the mean number detected in two technical replicates individually sampled at each read depth. (B) Normalized miRNA read counts (number of miRNA reads per 1 million human alignments) for each method. Colored bars represent the miRNA 5′-nucleotide. (C) Total miRNA species counts sorted by the identity of the 5′ end nucleotide. Both miRNA strands (miRNA-5p and miRNA-3p) are counted for this analysis. (D) Overlap of unique miRNAs identified in two combined replicates between CapTS-seq, TS-seq (uncapped) and SMARTer libraries. CapTS-seq detected more unique miRNAs. RPM, reads per million mapped reads.