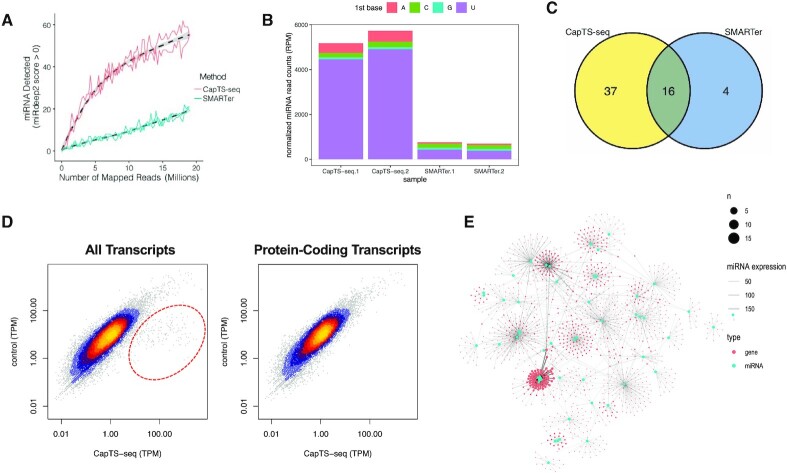
Figure 6.
CapTS-seq improves miRNAs detection in FFPE RNA and enables simultaneous transcriptome analysis. (A) Number of miRNAs detected with a mirdeep2 score >0 at various subsampled read depths. Datasets were randomly sampled with the number of reads increasing by 5000 in each sample. Values represent the mean number detected in two technical replicates individually sampled at each read depth. (B) Normalized miRNA read counts (number of miRNA reads per 1 million human alignments) for each method. Colored bars represent the miRNA 5′-nucleotide. (C) Overlap of unique miRNAs identified between CapTS-seq and SMARTer libraries. CapTS-seq detected more unique miRNAs. (D) Correlation of normalized read counts between CapTS-seq and a control RNA-seq library (NEBNext Ultra II). Left panel, all transcripts; right panel, protein-coding transcripts only. TPM, transcripts per kilobase million. TPM values were averaged between two technical replicates. Spearman's rank correlation coefficients for protein-coding reads (0.92) and for all transcripts (0.76). Reads within the red circle highlight small RNAs that are uniquely detected in CapTS-seq but not in a standard transcriptome RNA-seq library. (E) Interaction network of miRNAs (cyan dots) and their target genes (red dots) as detected in CapTS-seq libraries. The size of the dots represents the number of miRNAs that target this particular gene. The width of the connecting lines reflects the expression level of individual miRNAs.