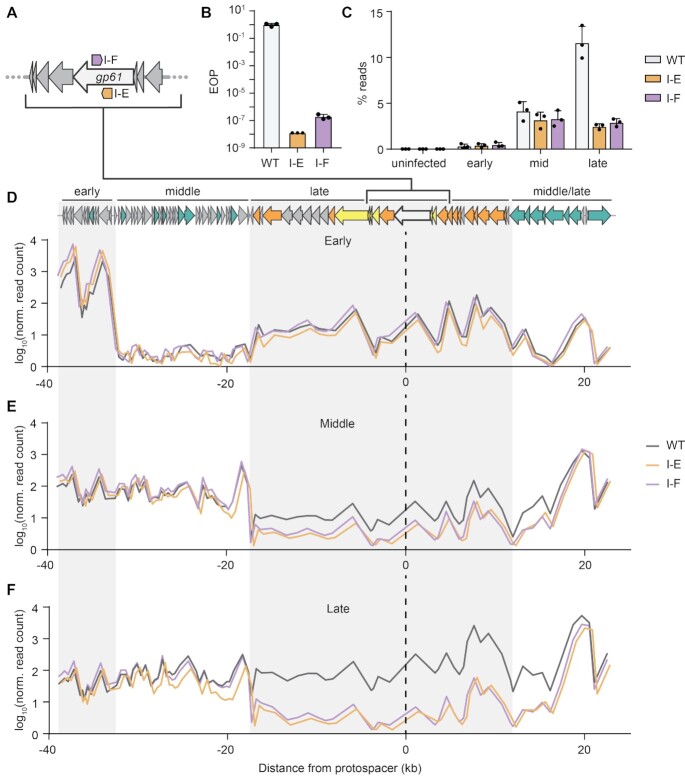
Figure 4.
Targeting by type I CRISPR-Cas systems affects transcription of phage genes during mid and late infection. (A) Schematic of type I-E and I-F anti-JS26 spacers targeting the tape measure gene (gp61). (B) EOP assay on strains carrying anti-JS26 spacers in the CRISPR native arrays CRISPR1 (type I-E, PCF524) and CRISPR2 (type I-F, PCF525). (C) Percentage of reads mapping to JS26 in strains without (WT, grey) or carrying type I-E (orange) or type I-F (purple) anti-phage spacers during early, middle and late stages of infection. Mean count of reads mapping to each gene across the phage genome during (D) early (E) mid and (F) late infection. In (D)–(F), read count is represented as mean of log10(x + 1) and distance from protospacer (PS) (black dashed line) was calculated from the midpoint of each gene. The small shift observed for the type I-F targeting strain is due to the distance between the location of the type I-E versus the type I-F protospacer. Grey and white boxes represent clusters of genes expressed during early, middle, middle/late and late stages of infection. In B and C, data are represented as mean of biological triplicates ± SD.