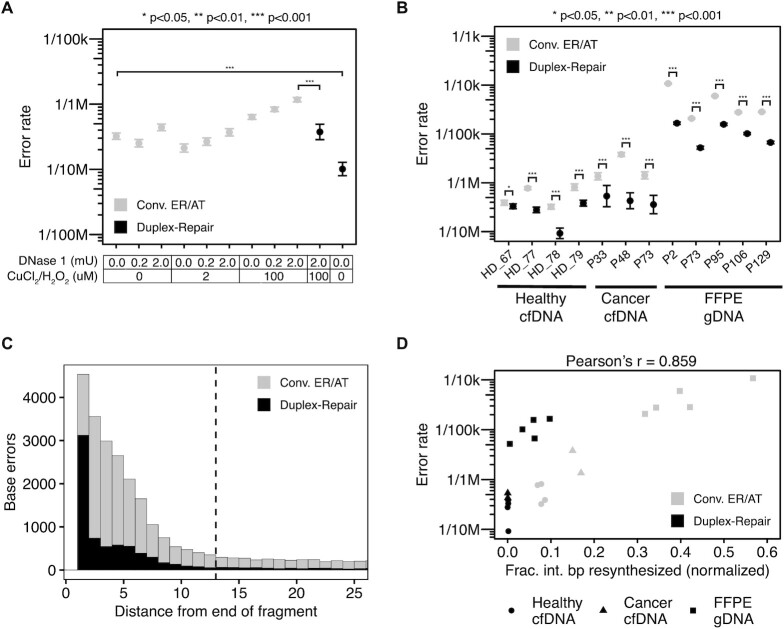
Figure 3.
Targeted panel sequencing of cfDNA and FFPE tumor biopsies. (A) Measured duplex sequencing error rates of HD_78 cfDNA damaged with varied concentrations of DNase I (to induce nicks) and CuCl2/H2O2 (to induce oxidative damage) and then repaired by using Duplex-Repair or conventional ER/AT (three replicates per condition). (B) Duplex sequencing error rates of four healthy cfDNA samples (three replicates per condition), three cancer patient cfDNA samples (one replicate per condition), and five cancer patient FFPE tumor biopsies (three replicates per condition) treated with conventional ER/AT or Duplex-Repair. (C) Aggregate mutant bases and their position relative to the end of the original duplex fragment. Dashed line represents the threshold of the interior of the fragment (12 bp). (D) Error rates from (B) compared to their corresponding estimates of interior base pair resynthesis fractions from Figure 2D. Pearson's correlation calculated for all data points.