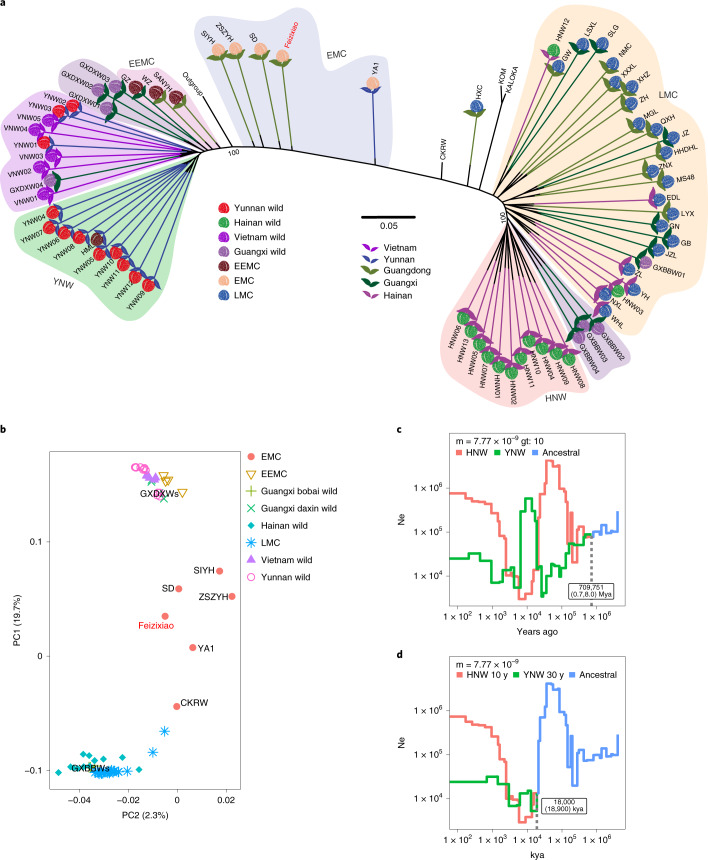
Fig. 2. Population analyses of resequenced lychee varieties.
a, A tree of all lychee accessions estimated based on high-quality SNPs. The black portion of each branch, representing the actual genetic distance, is extended (with colored lines) to better show the tree structure. b, PCA using all identified SNPs as markers closely recapitulates the tree in a by identifying two main clusters along PC1 (GXBBWs and LMC, versus GXDXWs) with intermediate cultivars arrayed in between (clustering with ‘Feizixiao’). c, The joint population history (blue line) of Yunnan wild (YNW, green line) and Hainan wild populations (HNW, red line) and their split time (dashed line) estimated using SMC++28. Different mutation rate-generation time combinations were used to obtain the ranges of possible split times. d, After correcting for inbreeding, the joint population history of Yunnan and Hainan wild populations show divergence at ~18 thousand years ago (kya) (Supplementary Note II).