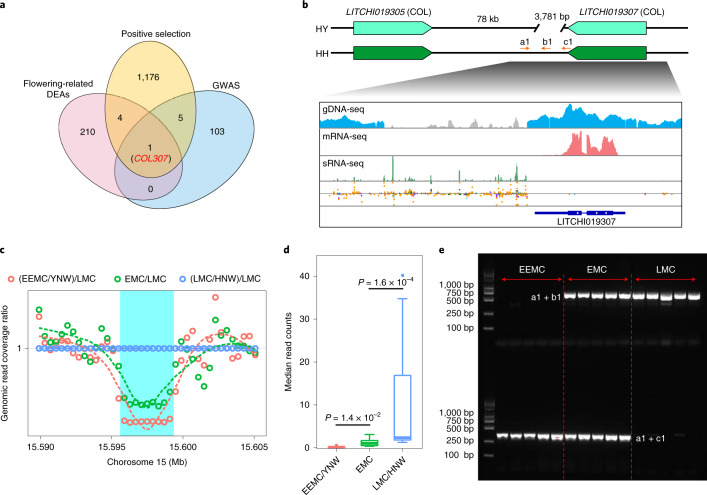
Fig. 6. A pair of COL genes associated with lychee fruit maturation time.
a, Venn diagram of positively selected genes, genes identified from the GWAS analysis and flowering-related DEA genes. b, Identification of a heterozygous 3.7-kb deletion downstream of the COL307 in the ‘Feizixiao’ genome. gDNA coverage over the deletion region is in gray. sRNA data are presented by either coverage in green or color-coded dots. Orange dots denote 24-nt siRNAs. Positions of PCR primers used for genotyping are labeled a1, b1, and c1. c, Lower coverage of genomic sequencing reads over the 3.7 kb region in EEMC/YNW and EMC cultivars suggests a deletion (light blue box). Open circles reflect average genomic coverage in 800 bp windows across chromosome 15. d, The coverage over the deletion region is significantly lower in the EEMC/YNW and EMC cultivars than in the LMC/HNW cultivars. P values were calculated with two-sided Student’s t-test. Numbers of accessions: n = 8 (EMC), n = 20 (EEMC/YNW) and n = 36 (LMC/HNW). Box plots show the median, box edges represent the 25th and 75th percentiles and whiskers represent the maximum and minimum data points within 1.5× interquartile range outside box edges. e, The 3.7-kb deletion can be used as a molecular marker to distinguish accessions from the EEMC, EMC and LMC cultivar groups. The PCR amplification was performed once.