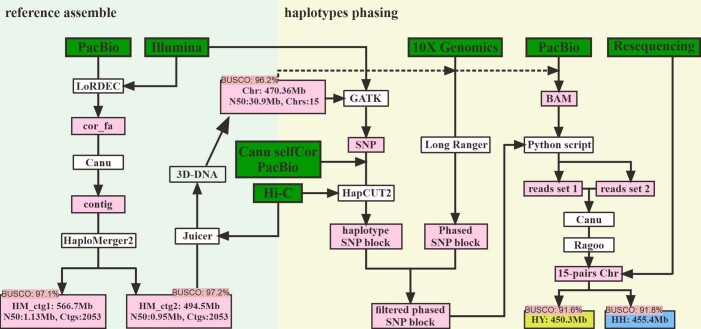
Extended Data Fig. 1. Workflow of genome assembly and haplotype phasing.
PacBio subreads were corrected using Illumina PE reads with LoRDEC, followed by assembly with the Canu software. Afterward, assembled contigs were separated into two contig collections using HaploMerger2, and one of the collections (HM_ctg2) with similar genome size to our flow cytometric estimation was used for the anchoring of assembled contigs into pseudochromosomes using Juicer and 3D-DNA based on Hi-C data. Finally, a reference genome, totaling 470 M with 15 chromosomes, was obtained. Haplotype phasing: Illumina reads were mapped against the reference genome and the resulting alignment file was used for variant calling following the GATK4 pipeline. Subsequently, hard-filter passing and biallelic SNPs (11.2 M) were kept for haplotype phasing using HapCUT2. Meanwhile, 10X Genomics reads were used for calling phased SNP blocks. SNPs (6.4 M) combined from the above two sets of SNP blocks were then retained for PacBio reads phasing using the approach first developed for the phasing of sex chromosomes (Zhang et al., 2020). After that, phased reads were de novo assembled with Canu independently, and contigs were linked with the help of the reference genome using RaGOO software. Eventually, 15 pairs of homologous pseudo-chromosomes were obtained and divided into two haplotypes, HY and HH, according to the mapping rates of resequencing data.