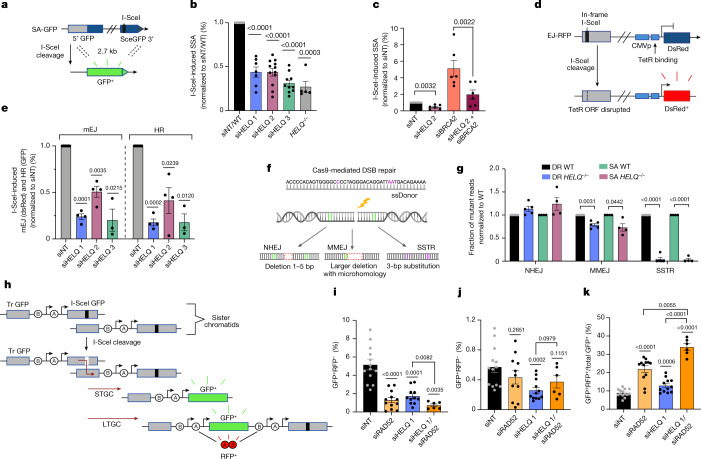
Fig. 4. HELQ functions in DSB repair, SSA and MMEJ.
a, Schematic of the SA-GFP reporter assay. b, I-SceI-induced SSA frequency in U2OS-SA HELQ–/– cells and WT cells treated with the indicated short interfering RNA (siRNA). n = 17 (non-targeting siRNA (siNT)/WT), n = 7 (siHELQ 1), n = 12 (siHELQ_2), n = 10 (siHELQ 3) and n = 5 (HELQ–/–) independent experiments. Data are mean ± s.e.m. c, SSA frequency in U2OS-SA cells treated with the indicated siRNA. n = 6 independent experiments. Data are mean ± s.e.m. d, Schematic of the EJ-RFP reporter assay. e, I-SceI-induced mutagenic end-joining (mEJ-RFP) and HR (DR-GFP) frequencies in U2OS-EJDR cells treated with the indicated siRNA. n = 7 (siNT), n = 4 (siHELQ 1), n = 2 (siHELQ 2) and n = 3 (siHELQ 3) independent experiments. Data are mean ± s.e.m. f, Schematic of Cas9-mediated DSB repair assay. g, Cas9-mediated DSB repair assay showing the frequencies of NHEJ, MMEJ and SSTR repair in HELQ–/– and WT U2OS-DR or U2OS-SA cells. n = 5 (U2OS-DR WT and DR HELQ–/–) and n = 4 (U2OS-SA and SA HELQ–/–) independent experiments. Data are mean ± s.e.m. h, Schematic of the RFP-SCR reporter assay. Tr GFP, truncated GFP. i, I-SceI-induced STGC frequency in U2OS-RFP-SCR cells treated with the indicated siRNA. n = 13 (siNT) n = 11 (siHELQ 1), n = 12 (siRAD52) and n = 6 (siHELQ 1/siRAD52) independent experiments. Data are mean ± s.e.m. j, LTGC frequency as shown in i. n = 13 (siNT), n = 12 (siHELQ 1), n = 11 (siRAD52) and n = 6 (siHELQ 1/siRAD52) independent experiments. Data are mean ± s.e.m. k, The ratio of LTGC/total GC from the experiments in i and j. n = 14 (siNT), n = 12 (siHELQ 1), n = 12 (siRAD52) and n = 6 (siHELQ 1/siRAD52) independent experiments. Data are mean ± s.e.m. For b, c, e, g, i–k, statistical analysis was performed using two-tailed paired t-tests as compared with siNT, unless otherwise indicated.