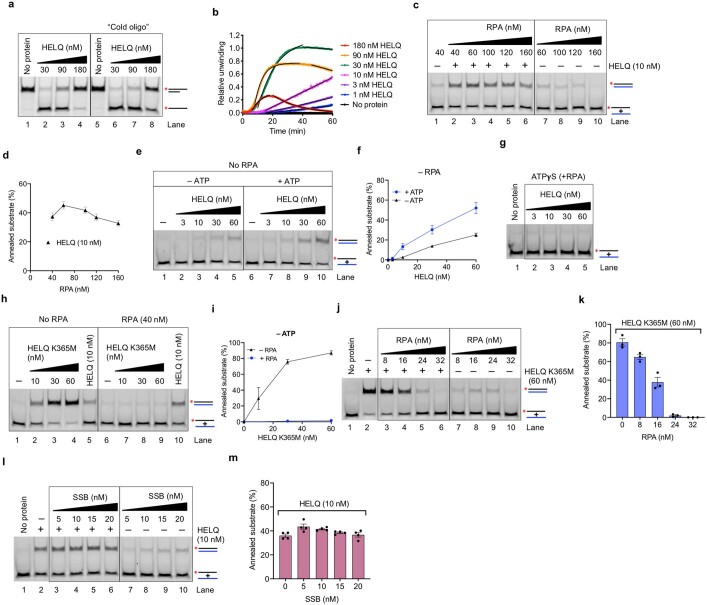
Extended Data Fig. 5. ATP is important for HELQ annealing activity in presence of RPA.
a, Representative gel of DNA unwinding assay of 3′-overhang with HELQ, in the presence and absence of “cold oligo” (25 nM) i.e., unlabelled oligo with identical DNA sequence as FITC-labelled oligo. b, Relative unwinding of 3′ overhang with indicated concentrations of HELQ as determined by quenching-based kinetic assay. n = 3 independent experiments; shaded area represents mean ± S.E.M.; black lines represent exponential or linear fits. c, Representative gel of DNA annealing assay with HELQ (10 nM) and indicated concentrations of RPA. The black and blue colours of substrate represent complementary DNA strands. The asterisk indicates the position of FITC labelling at 5′ end. The products were resolved on 10% native polyacrylamide gel. d, Quantification of experiments such as shown in c. n = 5 independent experiments; mean ± S.E.M. e, Representative gel of DNA annealing assay with indicated concentrations of HELQ in the absence and presence of ATP. f, Quantification of experiments such as shown in e. n= 3 independent experiments; mean ± S.E.M. g, Native gel showing DNA annealing assay with indicated concentrations of HELQ and RPA (40 nM) in the presence of ATPγS (n=2). h, Representative gel of DNA annealing assay with indicated concentrations of HELQ(K365M) in the absence and presence of RPA (40 nM). i, Quantification of experiments such as shown in h. n = 4 independent experiments; mean ± S.E.M. j, Representative gel of DNA annealing assay with HELQ(K365M) (60 nM) and indicated concentrations of RPA. k, Quantification of experiments such as shown in j. n = 3 independent experiments; mean ± S.E.M. l, Representative gel of DNA annealing assay with HELQ (10 nM) and various concentrations of SSB. m, Quantification of experiments such as shown in l. n = 4 independent experiments; mean ± S.E.M.