Figure 4. CX3CR1+Tstem-l have a distinct transcriptional signature.
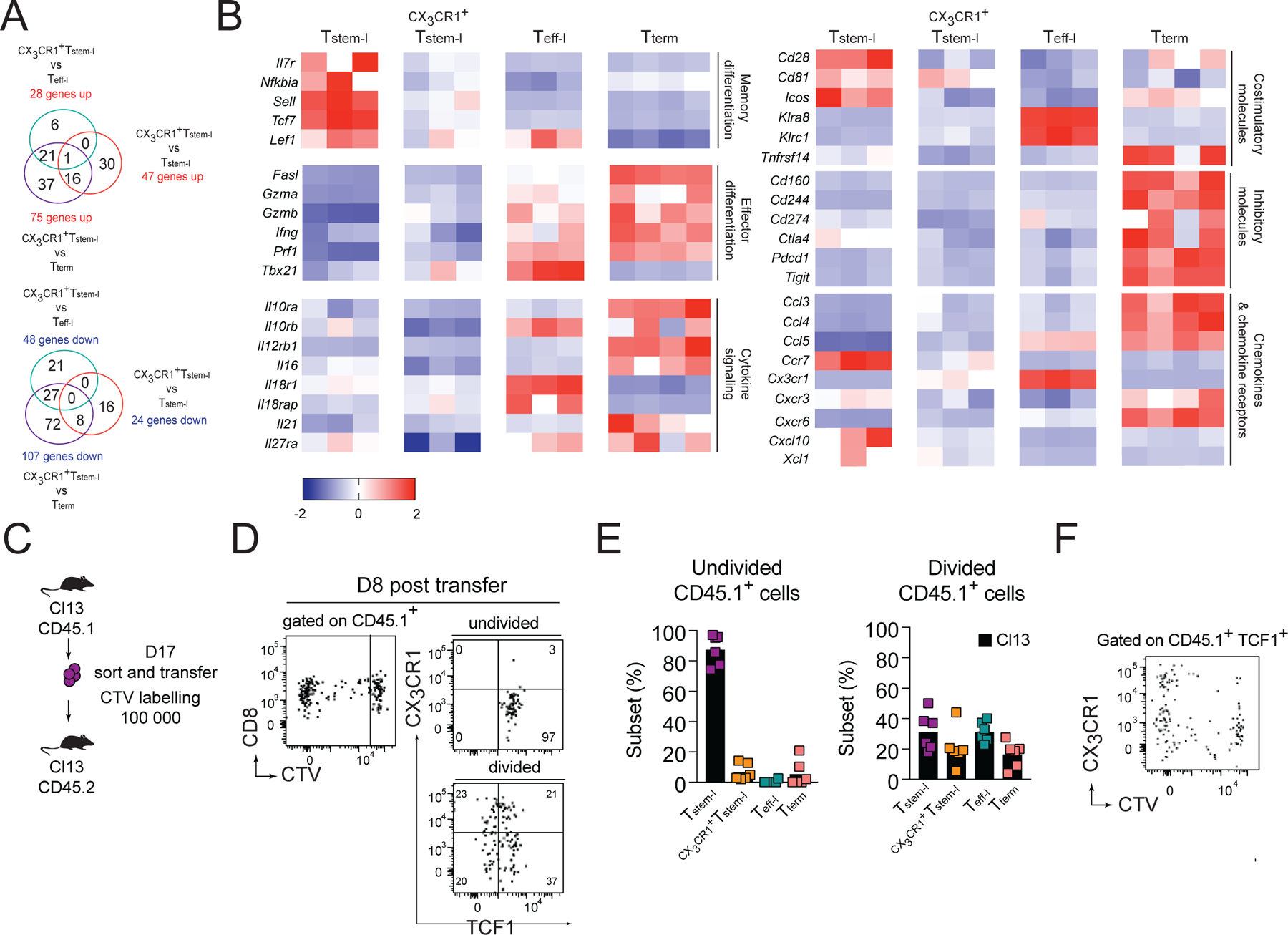
(A-B) Ly108, CX3CR1 and CD101 were used to distinguish and sort four T cell subsets from WT mice for analysis of differentially expressed genes using NanoString. (A) Number of differentially expressed genes between CX3CR1+Tstem-l and other T cell subsets. (B) Heatmap representing relative expression of selected differentially expressed genes between all subsets. (C-F) Congenically marked CX3CR1−Ly108+CD44+PD1+ CD8+ T cells (Tstem-l) were sorted from d17 Cl13-infected mice, labelled with cell tracker violet (CTV) and transferred into infection matched recipients. (C) Experimental scheme. (D) Proliferation and differentiation of Tstem-l in the spleen on d8 post-transfer. (E) Frequency of distinct subsets among undivided and divided cells. (F) Representative flow cytometry plot showing CX3CR1 and CTV expression on transferred cells that remained TCF1 positive. Data are from at least two independent experiments (n=3–4 per time point). Data in B were analyzed with the use of nSolver and multiple comparison test with Benjamini-Yekutieli False Discovery Rate method; *, P < 0.05; **, P < 0.01; ***, P < 0.001. See also Figure S2.
