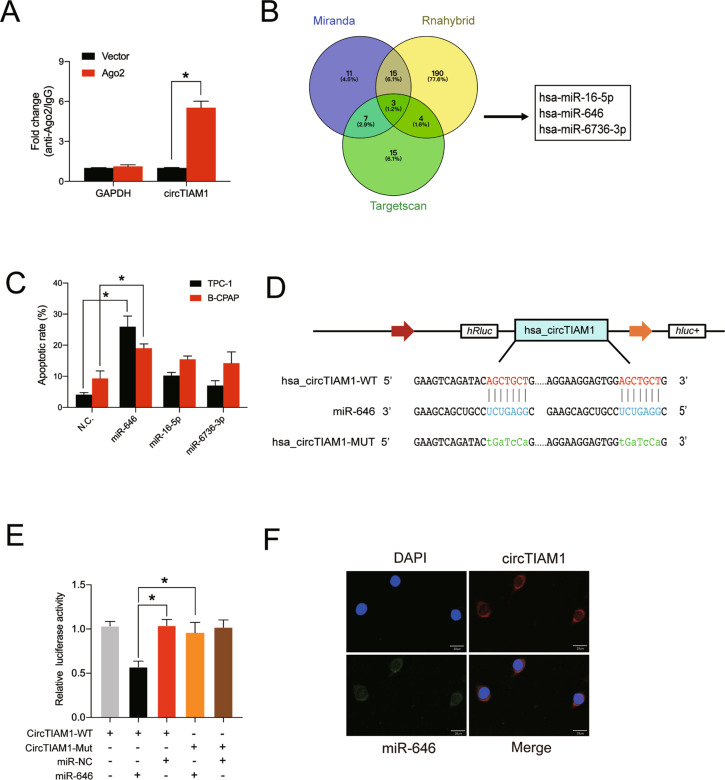
Fig. 3. CircTIAM1 serves as a sponge for miR-646 in PTC cells.
A Ago2 RIP assay was conducted to evaluate circTIAM1 pulled down by the anti-Ago2 antibody in B-CPAP cells transfected with Ago2 plasmid or vector. B Schematic diagram showing the potential microRNAs that bind to circTIAM1, revealed by the overlapping of the TargetScan, miRanda, and RNAhybrid databases. C TPC-1 and B-CPAP cells were transfected with target microRNA mimics or mimic NC, and the cells were collected and stained with Annexin V-FITC/PI after 48 h. The apoptotic rates were shown as mean ± S.D. D Schematic diagram of the binding sequence of circTIAM1 and miR-646. Mutated nucleotides of circTIAM1 are represented in lowercase letters. E Luciferase reporter assay was conducted to evaluate the luciferase activity of HEK-293T co-transfected with miR-646 mimics or NC and the luciferase reporter containing wild-type or mutated circTIAM1. F FISH revealed co-localization of circTIAM1 and miR-646 in TPC-1 cells. CircTIAM1 probes were labeled with Cy3, miR-646 probes were labeled with FAM, and DAPI was chosen to stain the nucleus (Scale bar = 20 μm). Data represent the mean ± SD. Three independent assays were performed (A&C) (*p < 0.05; Student’s t-test).