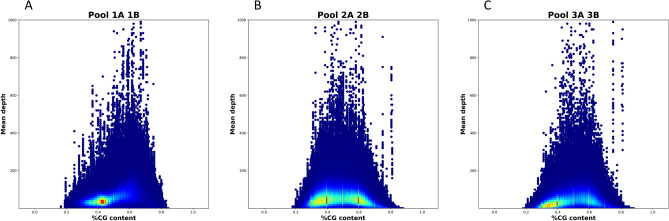
Figure 9.
Density plot of %GC Content vs Mean Depth for: (A) RSMU_exome + Agilent v6; (B) RSMU_exome + MGI v4; (C) MGIeasy capture + MGI v4. Here we show a 2D density plot of %GC Content vs Mean Depth parameters calculated by Picard HsMetrics v2.22.4. We obtained the data for this plot by merging all samples from the corresponding pools (1A + 1B, 2A + 2B, 3A + 3B) with Picard 2.22.4 . Density estimation was performed using 2D histograms. More specifically, we chose data points in a fixed rectangle (%GC Content ∈ (0;1) and Mean Depth ∈ (0;1000) ), then we split this rectangle into an evenly spaced grid of the size 200 × 100, and counted the number of data points in each cell of the grid. Finally, we normalized the grid to the (0,1) range and plotted it using "jet" colormap from the matplotlib library (https://matplotlib.org/).